Phylogenetic analysis based evolutionary study of 16S rRNA in known Pseudomonas sp
- PMID: 26664032
- PMCID: PMC4658646
- DOI: 10.6026/97320630011474
Phylogenetic analysis based evolutionary study of 16S rRNA in known Pseudomonas sp
Abstract
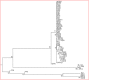
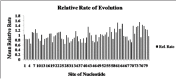
Molecular evolution analysis of 16S rRNA sequences of native Pseudomonas strains and different fluorescent pseudomonads were conducted on the basis of Molecular Evolutionary Genetics Analysis version 5.2 (MEGA5.2). Topological evaluations show common origin for native strains with other known strains with available sequences at GenBank database. Phylogenetic affiliation of different Pseudomonas sp based on 16S rRNA gene shows that molecular divergence contributes to the genetic diversity of Pseudomonas sp. Result indicate direct dynamic interactions with the rhizospheric pathogenic microbial community. The selection pressure acting on 16S rRNA gene was related to the nucleotide diversity of Pseudomonas sp in soil rhizosphere community among different agricultural crops. Besides, nucleotide diversity among the whole population was very low and tajima test statistic value (D) was also slightly positive (Tajima׳s test statistics D value 0.351). This data indicated increasing trends of infection of soil-borne pathogens under gangetic-alluvial regions of West Bengal due to high degree of nucleotide diversity with decreased population of plant growth promoting rhizobacteria like fluorescent Pseudomonads in soil.
Keywords: 16S rRNA; MEGA5.2; Molecular diversity; Positive selection; Pseudomonas sp.
Figures
References
-
- Elomari M, et al. Int J Syst Bacteriol. 1997;47:846. - PubMed
-
- Dabboussi F, et al. Res Microbiol. 1999;150:303. - PubMed
-
- Elomari M, et al. Int J Syst Bacteriol. 1996;46:1138. - PubMed
-
- Godfrey SAC, et al. J Appl Microbiol. 2001;91:412. - PubMed
-
- Behrendt U, et al. Int J Syst Evol Microbiol. 2007;57:979. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
