Ribosomal protein and biogenesis factors affect multiple steps during movement of the Saccharomyces cerevisiae Ty1 retrotransposon
- PMID: 26664557
- PMCID: PMC4673737
- DOI: 10.1186/s13100-015-0053-5
Ribosomal protein and biogenesis factors affect multiple steps during movement of the Saccharomyces cerevisiae Ty1 retrotransposon
Erratum in
-
Erratum to: ribosomal protein and biogenesis factors affect multiple steps during movement of the Saccharomyces cerevisiae Ty1 retrotransposon.Mob DNA. 2016 Feb 9;7:5. doi: 10.1186/s13100-016-0060-1. eCollection 2016. Mob DNA. 2016. PMID: 26865864 Free PMC article.
Abstract
Background: A large number of Saccharomyces cerevisiae cellular factors modulate the movement of the retrovirus-like transposon Ty1. Surprisingly, a significant number of chromosomal genes required for Ty1 transposition encode components of the translational machinery, including ribosomal proteins, ribosomal biogenesis factors, protein trafficking proteins and protein or RNA modification enzymes.
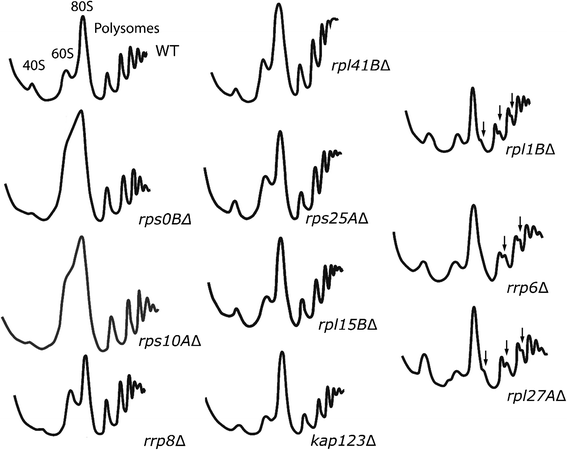
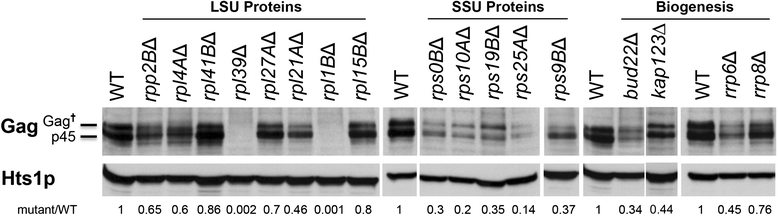
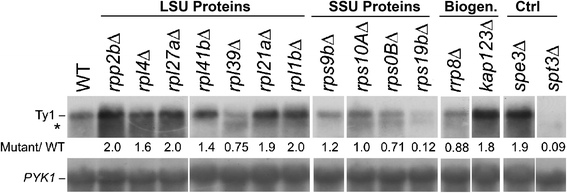
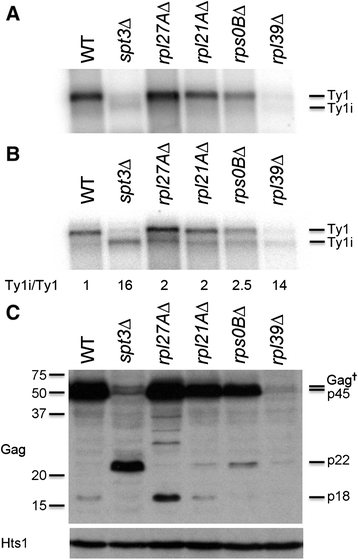
Results: To assess the mechanistic connection between Ty1 mobility and the translation machinery, we have determined the effect of these mutations on ribosome biogenesis and Ty1 transcriptional and post-transcriptional regulation. Lack of genes encoding ribosomal proteins or ribosome assembly factors causes reduced accumulation of the ribosomal subunit with which they are associated. In addition, these mutations cause decreased Ty1 + 1 programmed translational frameshifting, and reduced Gag protein accumulation despite at least normal levels of Ty1 mRNA. Several ribosome subunit mutations increase the level of both an internally initiated Ty1 transcript and its encoded truncated Gag-p22 protein, which inhibits transposition.
Conclusions: Together, our results suggest that this large class of cellular genes modulate Ty1 transposition through multiple pathways. The effects are largely post-transcriptional acting at a variety of levels that may include translation initiation, protein stability and subcellular protein localization.
Keywords: Host factors; Programmed frameshifting; Retrotransposition; Ribosomal protein insufficiency; Ribosome biogenesis.
Figures

Similar articles
-
Ribosome Biogenesis Modulates Ty1 Copy Number Control in Saccharomyces cerevisiae.Genetics. 2017 Dec;207(4):1441-1456. doi: 10.1534/genetics.117.300388. Epub 2017 Oct 18. Genetics. 2017. PMID: 29046400 Free PMC article.
-
Host co-factors of the retrovirus-like transposon Ty1.Mob DNA. 2012 Aug 2;3(1):12. doi: 10.1186/1759-8753-3-12. Mob DNA. 2012. PMID: 22856544 Free PMC article.
-
BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae.Genetics. 2010 Aug;185(4):1193-205. doi: 10.1534/genetics.110.119115. Epub 2010 May 24. Genetics. 2010. PMID: 20498295 Free PMC article.
-
A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces.Curr Genet. 2016 May;62(2):321-9. doi: 10.1007/s00294-015-0550-6. Epub 2015 Dec 9. Curr Genet. 2016. PMID: 26650614 Free PMC article. Review.
-
Happy together: the life and times of Ty retrotransposons and their hosts.Cytogenet Genome Res. 2005;110(1-4):70-90. doi: 10.1159/000084940. Cytogenet Genome Res. 2005. PMID: 16093660 Review.
Cited by
-
Erratum to: ribosomal protein and biogenesis factors affect multiple steps during movement of the Saccharomyces cerevisiae Ty1 retrotransposon.Mob DNA. 2016 Feb 9;7:5. doi: 10.1186/s13100-016-0060-1. eCollection 2016. Mob DNA. 2016. PMID: 26865864 Free PMC article.
-
Ribosome Biogenesis Modulates Ty1 Copy Number Control in Saccharomyces cerevisiae.Genetics. 2017 Dec;207(4):1441-1456. doi: 10.1534/genetics.117.300388. Epub 2017 Oct 18. Genetics. 2017. PMID: 29046400 Free PMC article.
-
Host factors that promote retrotransposon integration are similar in distantly related eukaryotes.PLoS Genet. 2017 Dec 12;13(12):e1006775. doi: 10.1371/journal.pgen.1006775. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29232693 Free PMC article.
-
Reliance of Host-Encoded Regulators of Retromobility on Ty1 Promoter Activity or Architecture.Front Mol Biosci. 2022 Jul 1;9:896215. doi: 10.3389/fmolb.2022.896215. eCollection 2022. Front Mol Biosci. 2022. PMID: 35847981 Free PMC article.
-
A nuclear pore sub-complex restricts the propagation of Ty retrotransposons by limiting their transcription.PLoS Genet. 2021 Nov 1;17(11):e1009889. doi: 10.1371/journal.pgen.1009889. eCollection 2021 Nov. PLoS Genet. 2021. PMID: 34723966 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

