Production of fatty acids in Ralstonia eutropha H16 by engineering β-oxidation and carbon storage
- PMID: 26664804
- PMCID: PMC4675107
- DOI: 10.7717/peerj.1468
Production of fatty acids in Ralstonia eutropha H16 by engineering β-oxidation and carbon storage
Abstract
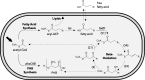
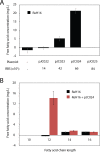
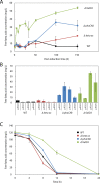
Ralstonia eutropha H16 is a facultatively autotrophic hydrogen-oxidizing bacterium capable of producing polyhydroxybutyrate (PHB)-based bioplastics. As PHB's physical properties may be improved by incorporation of medium-chain-length fatty acids (MCFAs), and MCFAs are valuable on their own as fuel and chemical intermediates, we engineered R. eutropha for MCFA production. Expression of UcFatB2, a medium-chain-length-specific acyl-ACP thioesterase, resulted in production of 14 mg/L laurate in wild-type R. eutropha. Total fatty acid production (22 mg/L) could be increased up to 2.5-fold by knocking out PHB synthesis, a major sink for acetyl-CoA, or by knocking out the acyl-CoA ligase fadD3, an entry point for fatty acids into β-oxidation. As ΔfadD3 mutants still consumed laurate, and because the R. eutropha genome is predicted to encode over 50 acyl-CoA ligases, we employed RNA-Seq to identify acyl-CoA ligases upregulated during growth on laurate. Knockouts of the three most highly upregulated acyl-CoA ligases increased fatty acid yield significantly, with one strain (ΔA2794) producing up to 62 mg/L free fatty acid. This study demonstrates that homologous β-oxidation systems can be rationally engineered to enhance fatty acid production, a strategy that may be employed to increase yield for a range of fuels, chemicals, and PHB derivatives in R. eutropha.
Keywords: -oxidation; Acyl-CoA ligase; Biofuel; Metabolic engineering; Ralstonia.
Conflict of interest statement
The authors declare there are no competing interests.
Figures

References
-
- Bi C, Su P, Muller J, Yeh Y-C, Chhabra S, Beller H, Singer S, Hillson N. Development of a broad-host synthetic biology toolbox for Ralstonia eutropha and its application to engineering hydrocarbon biofuel production. Microbial Cell Factories. 2013;12:107. doi: 10.1186/1475-2859-12-107. - DOI - PMC - PubMed
Further reading
-
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nature Biotechnology. 1983;1:784–791. doi: 10.1038/nbt1183-784. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

