IRES-dependent translated genes in fungi: computational prediction, phylogenetic conservation and functional association
- PMID: 26666532
- PMCID: PMC4678720
- DOI: 10.1186/s12864-015-2266-x
IRES-dependent translated genes in fungi: computational prediction, phylogenetic conservation and functional association
Abstract
Background: The initiation of translation via cellular internal ribosome entry sites plays an important role in the stress response and certain physiological conditions in which canonical cap-dependent translation initiation is compromised. Currently, only a limited number of these regulatory elements have been experimentally identified. Notably, cellular internal ribosome entry sites lack conservation of both the primary sequence and mRNA secondary structure, rendering their identification difficult. Despite their biological importance, the currently available computational strategies to predict them have had limited success. We developed a bioinformatic method based on a support vector machine for the prediction of internal ribosome entry sites in fungi using the 5'-UTR sequences of 20 non-redundant fungal organisms. Additionally, we performed a comparative analysis and characterization of the functional relationships among the gene products predicted to be translated by this cap-independent mechanism.
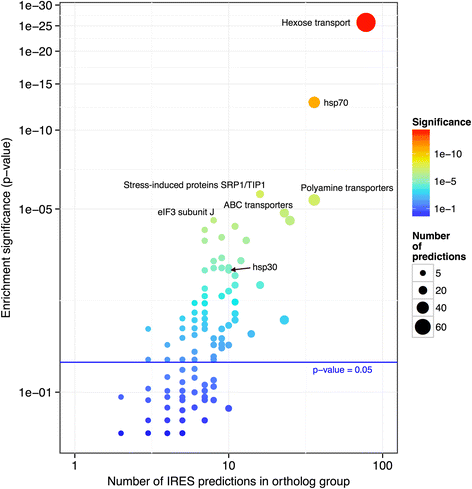
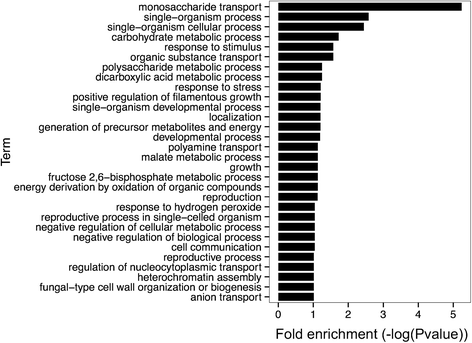
Results: Using our method, we predicted 6,532 internal ribosome entry sites in 20 non-redundant fungal organisms. Some orthologous groups were enriched with our positive predictions. This is the case of the HSP70 chaperone family, which remarkably has two verified internal ribosome entry sites, one in humans and the other in flies. A second example is the orthologous group of the eIF4G repression protein Sbp1p, which has two homologous genes known to be translated by this cap-independent mechanism, one in mice and the other in yeast. These examples emphasize the wide conservation of these regulatory elements as a result of selective pressure. In addition, we performed a protein-protein interaction network characterization of the gene products of our positive predictions using Saccharomyces cerevisiae as a model, which revealed a highly connected and modular topology, suggesting a functional association. A remarkable example of this functional association is our prediction of internal ribosome entry sites elements in three components of the RNA polymerase II mediator complex.
Conclusions: We developed a method for the prediction of cellular internal ribosome entry sites that may guide experimental and bioinformatic analyses to increase our understanding of protein translation regulation. Our analysis suggests that fungi show evolutionary conservation and functional association of proteins translated by this cap-independent mechanism.
Figures


Similar articles
-
Deconstructing internal ribosome entry site elements: an update of structural motifs and functional divergences.Open Biol. 2018 Nov 28;8(11):180155. doi: 10.1098/rsob.180155. Open Biol. 2018. PMID: 30487301 Free PMC article. Review.
-
IRESpy: an XGBoost model for prediction of internal ribosome entry sites.BMC Bioinformatics. 2019 Jul 30;20(1):409. doi: 10.1186/s12859-019-2999-7. BMC Bioinformatics. 2019. PMID: 31362694 Free PMC article.
-
IRESPred: Web Server for Prediction of Cellular and Viral Internal Ribosome Entry Site (IRES).Sci Rep. 2016 Jun 6;6:27436. doi: 10.1038/srep27436. Sci Rep. 2016. PMID: 27264539 Free PMC article.
-
Internal Initiation of Translation of mRNA in the Methylotrophic Yeast Hansenula polymorpha.Biochemistry (Mosc). 2016 May;81(5):521-9. doi: 10.1134/S0006297916050096. Biochemistry (Mosc). 2016. PMID: 27297902
-
Targeting internal ribosome entry site (IRES)-mediated translation to block hepatitis C and other RNA viruses.FEMS Microbiol Lett. 2004 May 15;234(2):189-99. doi: 10.1016/j.femsle.2004.03.045. FEMS Microbiol Lett. 2004. PMID: 15135522 Review.
Cited by
-
Deconstructing internal ribosome entry site elements: an update of structural motifs and functional divergences.Open Biol. 2018 Nov 28;8(11):180155. doi: 10.1098/rsob.180155. Open Biol. 2018. PMID: 30487301 Free PMC article. Review.
-
CNBP Homologues Gis2 and Znf9 Interact with a Putative G-Quadruplex-Forming 3' Untranslated Region, Altering Polysome Association and Stress Tolerance in Cryptococcus neoformans.mSphere. 2018 Aug 8;3(4):e00201-18. doi: 10.1128/mSphere.00201-18. mSphere. 2018. PMID: 30089646 Free PMC article.
-
RNA-Binding Proteins as Regulators of Internal Initiation of Viral mRNA Translation.Viruses. 2022 Jan 19;14(2):188. doi: 10.3390/v14020188. Viruses. 2022. PMID: 35215780 Free PMC article. Review.
-
First Evidence for Internal Ribosomal Entry Sites in Diverse Fungal Virus Genomes.mBio. 2018 Mar 20;9(2):e02350-17. doi: 10.1128/mBio.02350-17. mBio. 2018. PMID: 29559577 Free PMC article.
-
Siblings or doppelgängers? Deciphering the evolution of structured cis-regulatory RNAs beyond homology.Biochem Soc Trans. 2020 Oct 30;48(5):1941-1951. doi: 10.1042/BST20191060. Biochem Soc Trans. 2020. PMID: 32869842 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

