Effects of Stress, Reactive Oxygen Species, and the SOS Response on De Novo Acquisition of Antibiotic Resistance in Escherichia coli
- PMID: 26666928
- PMCID: PMC4776001
- DOI: 10.1128/AAC.02684-15
Effects of Stress, Reactive Oxygen Species, and the SOS Response on De Novo Acquisition of Antibiotic Resistance in Escherichia coli
Abstract
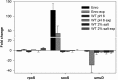
Strategies to prevent the development of antibiotic resistance in bacteria are needed to reduce the threat of infectious diseases to human health. The de novo acquisition of resistance due to mutations and/or phenotypic adaptation occurs rapidly as a result of interactions of gene expression and mutations (N. Handel, J. M. Schuurmans, Y. Feng, S. Brul, and B. H. Ter Kuile, Antimicrob Agents Chemother 58:4371-4379, 2014, http://dx.doi.org/10.1128/AAC.02892-14). In this study, the contribution of several individual genes to the de novo acquisition of antibiotic resistance in Escherichia coli was investigated using mutants with deletions of genes known to be involved in antibiotic resistance. The results indicate that recA, vital for the SOS response, plays a crucial role in the development of antibiotic resistance. Likewise, deletion of global transcriptional regulators, such as gadE or soxS, involved in pH homeostasis and superoxide removal, respectively, can slow the acquisition of resistance to a degree depending on the antibiotic. Deletion of the transcriptional regulator soxS, involved in superoxide removal, slowed the acquisition of resistance to enrofloxacin. Acquisition of resistance occurred at a lower rate in the presence of a second stress factor, such as a lowered pH or increased salt concentration, than in the presence of optimal growth conditions. The overall outcome suggests that a central cellular mechanism is crucial for the development of resistance and that genes involved in the regulation of transcription play an essential role. The actual cellular response, however, depends on the class of antibiotic in combination with environmental conditions.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

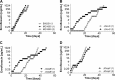



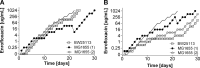
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

