Two-hierarchical nonnegative matrix factorization distinguishing the fluorescent targets from autofluorescence for fluorescence imaging
- PMID: 26667020
- PMCID: PMC4678484
- DOI: 10.1186/s12938-015-0107-4
Two-hierarchical nonnegative matrix factorization distinguishing the fluorescent targets from autofluorescence for fluorescence imaging
Abstract
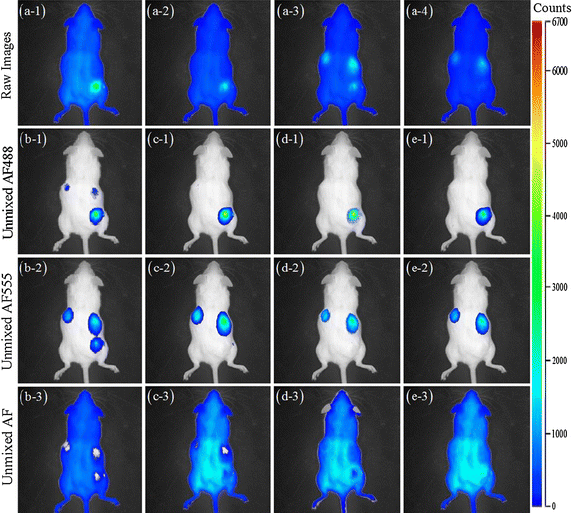
Background: Nonnegative matrix factorization (NMF) has been used in blind fluorescence unmixing for multispectral in-vivo fluorescence imaging, which decomposes a mixed source data into a set of constituent fluorescence spectra and corresponding concentrations. However, most classical NMF algorithms have ill convergence problems and they always fail to unmix multiple fluorescent targets from background autofluorescence for the sparse acquisition of multispectral fluorescence imaging, which introduces incomplete measurements and severe discontinuities in multispectral fluorescence emissions across the multiple spectral bands.
Methods: Observing the spatial distinction between the diffusive autofluorescence and the sparse fluorescent targets, we propose to separate the mixed sparse multispectral data into equality constrained two-hierarchical updating within NMF framework by dividing the concentration matrix of entire endmembers into two hierarchies: the fluorescence targets and the background autofluorescence. Specifically, when updating concentrations of multiple fluorescent targets in the two-hierarchical NMF, we assume that the concentration of autofluorescence is fixed and known, and vice versa. Furthermore, a sparsity constraint is imposed on the concentration matrix components of fluorescence targets only.
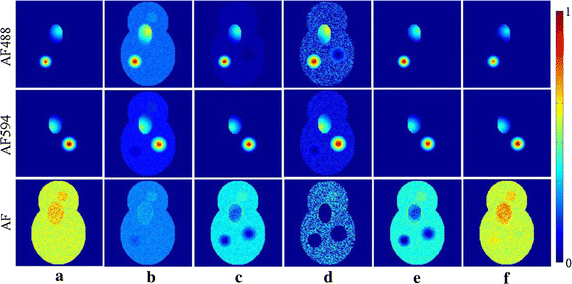
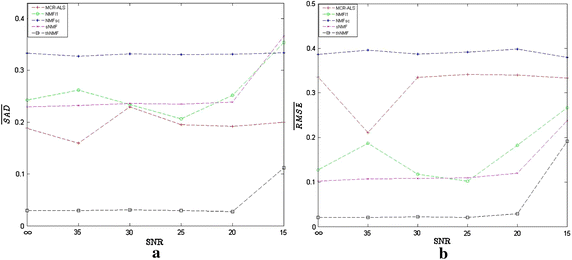
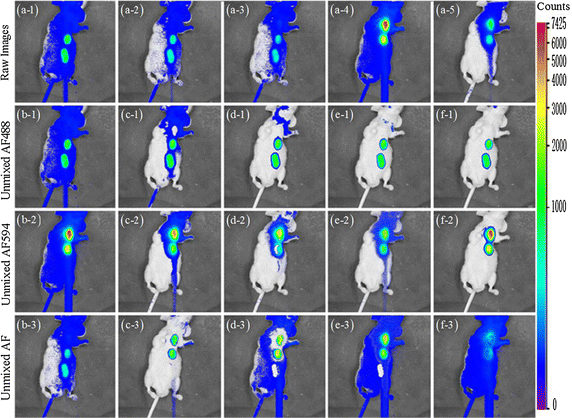
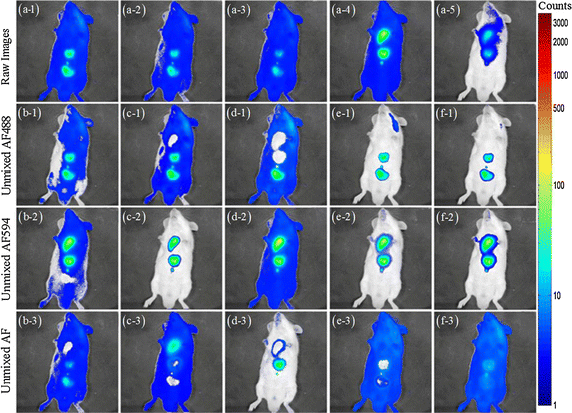
Results: Synthetic data sets, in vivo fluorescence imaging data are employed to demonstrate and validate the performance of our approach. The proposed algorithm can achieve more satisfying results of spectral unmixing and autofluorescence removal compared to other state-of-the-art methods, especially for the sparse multispectral fluorescence imaging.
Conclusions: The proposed algorithm can successfully tackle the sparse acquisition and ill-posed problems in the NMF-based fluorescence unmixing through equality constraint along with partial sparsity constraint during two-hierarchical NMF optimization, at which fixing sparsity constrained target fluorescence can make the update of autofluorescence as accurate as possible and vice versa.
Figures


Similar articles
-
Blind spectral unmixing based on sparse nonnegative matrix factorization.IEEE Trans Image Process. 2011 Apr;20(4):1112-25. doi: 10.1109/TIP.2010.2081678. Epub 2010 Sep 30. IEEE Trans Image Process. 2011. PMID: 20889432
-
Autofluorescence removal by non-negative matrix factorization.IEEE Trans Image Process. 2011 Apr;20(4):1085-93. doi: 10.1109/TIP.2010.2079810. Epub 2010 Sep 30. IEEE Trans Image Process. 2011. PMID: 20889433
-
In vivo fluorescence spectra unmixing and autofluorescence removal by sparse nonnegative matrix factorization.IEEE Trans Biomed Eng. 2011 Sep;58(9):2554-65. doi: 10.1109/TBME.2011.2159382. Epub 2011 Jun 13. IEEE Trans Biomed Eng. 2011. PMID: 21672672
-
Spectral Unmixing Methods and Tools for the Detection and Quantitation of Collagen and Other Macromolecules in Tissue Specimens.Methods Mol Biol. 2017;1627:491-509. doi: 10.1007/978-1-4939-7113-8_30. Methods Mol Biol. 2017. PMID: 28836220 Review.
-
A review of attenuation correction techniques for tissue fluorescence.J R Soc Interface. 2006 Feb 22;3(6):1-13. doi: 10.1098/rsif.2005.0066. J R Soc Interface. 2006. PMID: 16849213 Free PMC article. Review.
Cited by
-
Live-cell fluorescence spectral imaging as a data science challenge.Biophys Rev. 2022 Mar 23;14(2):579-597. doi: 10.1007/s12551-022-00941-x. eCollection 2022 Apr. Biophys Rev. 2022. PMID: 35528031 Free PMC article. Review.
-
Semi-blind sparse affine spectral unmixing of autofluorescence-contaminated micrographs.Bioinformatics. 2020 Feb 1;36(3):910-917. doi: 10.1093/bioinformatics/btz674. Bioinformatics. 2020. PMID: 31504202 Free PMC article.
-
Robust self-supervised denoising of voltage imaging data using CellMincer.bioRxiv [Preprint]. 2024 Apr 15:2024.04.12.589298. doi: 10.1101/2024.04.12.589298. bioRxiv. 2024. Update in: Npj Imaging. 2024;2(1):51. doi: 10.1038/s44303-024-00055-x. PMID: 38659950 Free PMC article. Updated. Preprint.
-
Robust self-supervised denoising of voltage imaging data using CellMincer.Npj Imaging. 2024;2(1):51. doi: 10.1038/s44303-024-00055-x. Epub 2024 Dec 4. Npj Imaging. 2024. PMID: 39649342 Free PMC article.
-
Robust blind spectral unmixing for fluorescence microscopy using unsupervised learning.PLoS One. 2019 Dec 2;14(12):e0225410. doi: 10.1371/journal.pone.0225410. eCollection 2019. PLoS One. 2019. PMID: 31790435 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

