Snf1-Dependent Transcription Confers Glucose-Induced Decay upon the mRNA Product
- PMID: 26667037
- PMCID: PMC4751693
- DOI: 10.1128/MCB.00436-15
Snf1-Dependent Transcription Confers Glucose-Induced Decay upon the mRNA Product
Abstract
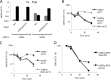
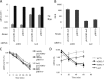
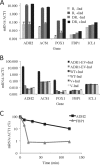
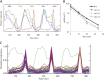
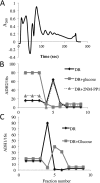
In the yeast Saccharomyces cerevisiae, the switch from respiratory metabolism to fermentation causes rapid decay of transcripts encoding proteins uniquely required for aerobic metabolism. Snf1, the yeast ortholog of AMP-activated protein kinase, has been implicated in this process because inhibiting Snf1 mimics the addition of glucose. In this study, we show that the SNF1-dependent ADH2 promoter, or just the major transcription factor binding site, is sufficient to confer glucose-induced mRNA decay upon heterologous transcripts. SNF1-independent expression from the ADH2 promoter prevented glucose-induced mRNA decay without altering the start site of transcription. SNF1-dependent transcripts are enriched for the binding motif of the RNA binding protein Vts1, an important mediator of mRNA decay and mRNA repression whose expression is correlated with decreased abundance of SNF1-dependent transcripts during the yeast metabolic cycle. However, deletion of VTS1 did not slow the rate of glucose-induced mRNA decay. ADH2 mRNA rapidly dissociated from polysomes after glucose repletion, and sequences bound by RNA binding proteins were enriched in the transcripts from repressed cells. Inhibiting the protein kinase A pathway did not affect glucose-induced decay of ADH2 mRNA. Our results suggest that Snf1 may influence mRNA stability by altering the recruitment activity of the transcription factor Adr1.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures




Similar articles
-
The AMP-activated protein kinase Snf1 regulates transcription factor binding, RNA polymerase II activity, and mRNA stability of glucose-repressed genes in Saccharomyces cerevisiae.J Biol Chem. 2012 Aug 17;287(34):29021-34. doi: 10.1074/jbc.M112.380147. Epub 2012 Jul 2. J Biol Chem. 2012. PMID: 22761425 Free PMC article.
-
Snf1-dependent and Snf1-independent pathways of constitutive ADH2 expression in Saccharomyces cerevisiae.Genetics. 2006 Apr;172(4):2123-38. doi: 10.1534/genetics.105.048231. Epub 2006 Jan 16. Genetics. 2006. PMID: 16415371 Free PMC article.
-
Snf1-independent, glucose-resistant transcription of Adr1-dependent genes in a mediator mutant of Saccharomyces cerevisiae.Mol Microbiol. 2009 Oct;74(2):364-83. doi: 10.1111/j.1365-2958.2009.06866.x. Epub 2009 Sep 2. Mol Microbiol. 2009. PMID: 19732343
-
Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae.Curr Genet. 2003 Jun;43(3):139-60. doi: 10.1007/s00294-003-0381-8. Epub 2003 Apr 25. Curr Genet. 2003. PMID: 12715202 Review.
-
Coupling mRNA synthesis and decay.Mol Cell Biol. 2014 Nov 15;34(22):4078-87. doi: 10.1128/MCB.00535-14. Epub 2014 Aug 25. Mol Cell Biol. 2014. PMID: 25154419 Free PMC article. Review.
Cited by
-
Reshaping the yeast galactose regulon via GPCR signaling cascade.Cell Rep Methods. 2023 Dec 18;3(12):100647. doi: 10.1016/j.crmeth.2023.100647. Epub 2023 Nov 20. Cell Rep Methods. 2023. PMID: 37989311 Free PMC article.
-
The interplay between transcription and mRNA degradation in Saccharomyces cerevisiae.Microb Cell. 2017 Jul 3;4(7):212-228. doi: 10.15698/mic2017.07.580. Microb Cell. 2017. PMID: 28706937 Free PMC article. Review.
-
Multilevel regulation of an α-arrestin by glucose depletion controls hexose transporter endocytosis.J Cell Biol. 2017 Jun 5;216(6):1811-1831. doi: 10.1083/jcb.201610094. Epub 2017 May 3. J Cell Biol. 2017. PMID: 28468835 Free PMC article.
-
Possible regulatory network and associated pathways governing the expression of ADH2 in Saccharomyces cerevisiae.Curr Genet. 2025 Aug 18;71(1):15. doi: 10.1007/s00294-025-01321-0. Curr Genet. 2025. PMID: 40824391 Review.
-
Promoter architecture determines cotranslational regulation of mRNA.Genome Res. 2018 Apr;28(4):509-518. doi: 10.1101/gr.230458.117. Epub 2018 Mar 22. Genome Res. 2018. PMID: 29567675 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
