The administration of intranasal live attenuated influenza vaccine induces changes in the nasal microbiota and nasal epithelium gene expression profiles
- PMID: 26667497
- PMCID: PMC4678663
- DOI: 10.1186/s40168-015-0133-2
The administration of intranasal live attenuated influenza vaccine induces changes in the nasal microbiota and nasal epithelium gene expression profiles
Abstract
Background: Viral infections such as influenza have been shown to predispose hosts to increased colonization of the respiratory tract by pathogenic bacteria and secondary bacterial pneumonia. To examine how viral infections and host antiviral immune responses alter the upper respiratory microbiota, we analyzed nasal bacterial composition by 16S ribosomal RNA (rRNA) gene sequencing in healthy adults at baseline and at 1 to 2 weeks and 4 to 6 weeks following instillation of live attenuated influenza vaccine or intranasal sterile saline. A subset of these samples was submitted for microarray host gene expression profiling.
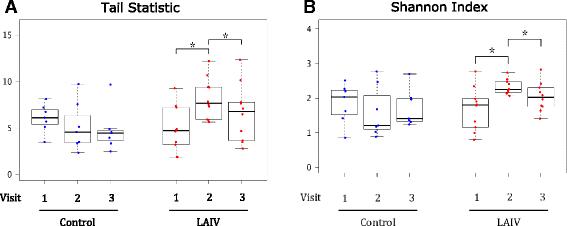
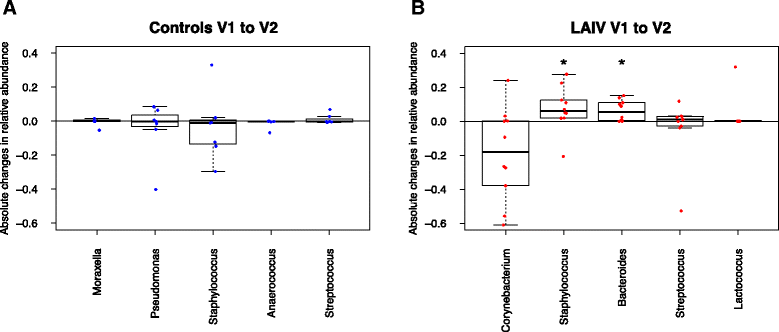
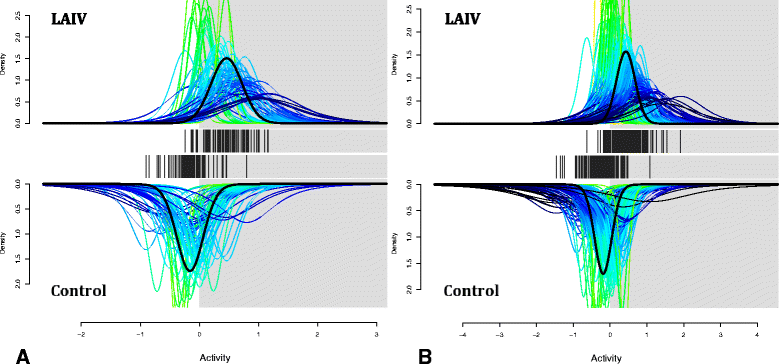
Results: We found that live attenuated influenza vaccination led to significant changes in microbial community structure, diversity, and core taxonomic membership as well as increases in the relative abundances of Staphylococcus and Bacteroides genera (both p < 0.05). Hypergeometric testing for the enrichment of gene ontology terms in the vaccinated group reflected a robust up-regulation of type I and type II interferon-stimulated genes in the vaccinated group relative to controls. Translational murine studies showed that poly I:C administration did in fact permit greater nasal Staphylococcus aureus persistence, a response absent in interferon alpha/beta receptor deficient mice.
Conclusions: Collectively, our findings demonstrate that although the human nasal bacterial community is heterogeneous and typically individually robust, activation of a type I interferon (IFN)-mediated antiviral response may foster the disproportionate emergence of potentially pathogenic species such as S. aureus.
Trial registration: This study was registered with Clinicaltrials.gov on 11/3/15, NCT02597647 .
Figures




References
-
- Louie J, Jean C, Chen T, Park S, Ueki R, Harper T, et al. Bacterial coinfections in lung tissue specimens from fatal cases of 2009 pandemic influenza A (H1N1)-United States, May-August 2009. Morb Mortal Wkly Rep. 2009;58(38):1071–4. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

