Mining candidate genes associated with powdery mildew resistance in cucumber via super-BSA by specific length amplified fragment (SLAF) sequencing
- PMID: 26668009
- PMCID: PMC4677437
- DOI: 10.1186/s12864-015-2041-z
Mining candidate genes associated with powdery mildew resistance in cucumber via super-BSA by specific length amplified fragment (SLAF) sequencing
Abstract
Background: Powdery mildew (PM) is the most common fungal disease of cucumber and other cucurbit crops, while breeding the PM-resistant materials is the effective way to defense this disease, and the recent development of modern genetics and genomics make us aware of that studying the resistance genes is the essential way to breed the PM high-resistance plant. With the ever increasing throughput of next-generation sequencing (NGS), the development of specific length amplified fragment sequencing (SLAF-seq) as a high-resolution strategy for large-scale de novo SNP discovery is gradually applied for functional gene mining. Here we combined the bulked segregant analysis (BSA) with SLAF-seq to identify candidate genes associated with PM resistance in cucumber.
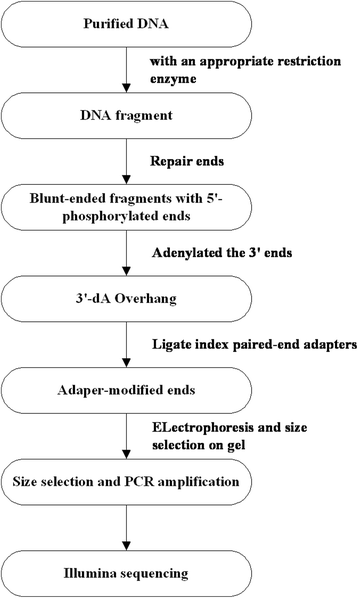
Methods: A segregating population comprising 251 F2 individuals was developed using H136 (female parent) as susceptible parent and BK2 (male parent) as resistance donor. After PMR test, total genomic DNA was prepared from each plant. Systemic genomic analysis of the GC content, repeat sequence, etc. was carried out by prediction software SLAF_Predict to establish condition to ensure the uniformity and density of the molecular markers. After samples were gel purified, SLAFs were generated at Biomarker Technologies Corporation in Beijing. Based on SLAF tags and the PMR test result, the hot region were annotated.
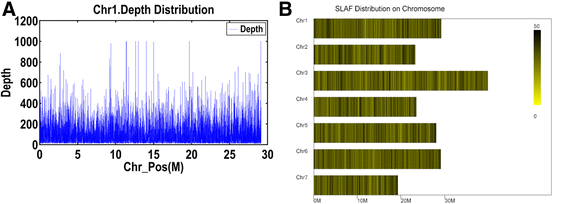
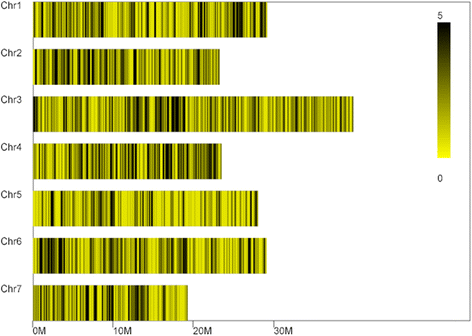
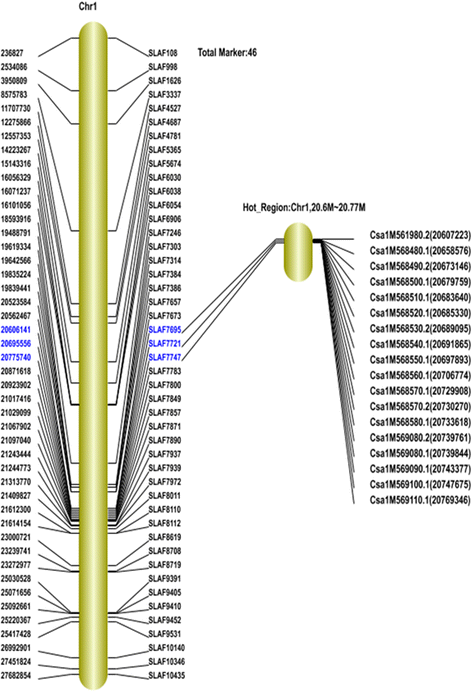
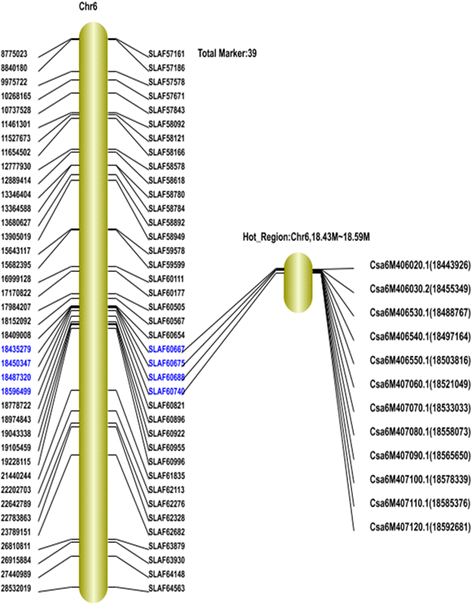
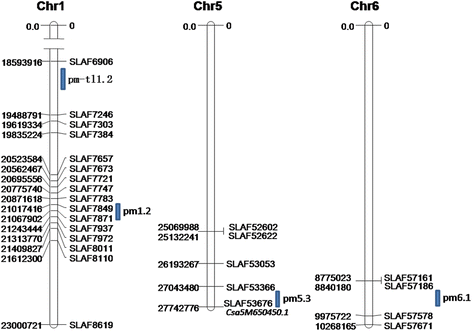
Results: A total of 73,100 high-quality SLAF tags with an average depth of 99.11× were sequenced. Among these, 5,355 polymorphic tags were identified with a polymorphism rate of 7.34 %, including 7.09 % SNPs and other polymorphism types. Finally, 140 associated SLAFs were identified, and two main Hot Regions were detected on chromosome 1 and 6, which contained five genes invovled in defense response, toxin metabolism, cell stress response, and injury response in cucumber.
Conclusions: Associated markers identified by super-BSA in this study, could not only speed up the study of the PMR genes, but also provide a feasible solution for breeding the marker-assisted PMR cucumber. Moreover, this study could also be extended to any other species with reference genome.
Figures



Similar articles
-
Candidate powdery mildew resistance gene in wheat landrace cultivar Hongyoumai discovered using SLAF and BSR-seq.BMC Plant Biol. 2022 Feb 23;22(1):83. doi: 10.1186/s12870-022-03448-5. BMC Plant Biol. 2022. PMID: 35196978 Free PMC article.
-
An SNP-based saturated genetic map and QTL analysis of fruit-related traits in cucumber using specific-length amplified fragment (SLAF) sequencing.BMC Genomics. 2014 Dec 22;15(1):1158. doi: 10.1186/1471-2164-15-1158. BMC Genomics. 2014. PMID: 25534138 Free PMC article.
-
Physical mapping and candidate gene prediction of fertility restorer gene of cytoplasmic male sterility in cotton.BMC Genomics. 2018 Jan 2;19(1):6. doi: 10.1186/s12864-017-4406-y. BMC Genomics. 2018. PMID: 29295711 Free PMC article.
-
Next generation sequencing and omics in cucumber (Cucumis sativus L.) breeding directed research.Plant Sci. 2016 Jan;242:77-88. doi: 10.1016/j.plantsci.2015.07.025. Epub 2015 Jul 31. Plant Sci. 2016. PMID: 26566826 Review.
-
Bulked sample analysis in genetics, genomics and crop improvement.Plant Biotechnol J. 2016 Oct;14(10):1941-55. doi: 10.1111/pbi.12559. Epub 2016 Apr 28. Plant Biotechnol J. 2016. PMID: 26990124 Free PMC article. Review.
Cited by
-
Cloning and Characterization of TaTGW-7A Gene Associated with Grain Weight in Wheat via SLAF-seq-BSA.Front Plant Sci. 2016 Dec 20;7:1902. doi: 10.3389/fpls.2016.01902. eCollection 2016. Front Plant Sci. 2016. PMID: 28066462 Free PMC article.
-
Genome-Wide Association Study of Major Agronomic Traits Related to Domestication in Peanut.Front Plant Sci. 2017 Sep 26;8:1611. doi: 10.3389/fpls.2017.01611. eCollection 2017. Front Plant Sci. 2017. PMID: 29018458 Free PMC article.
-
Fine mapping and molecular marker development of the Fs gene controlling fruit spines in spinach (Spinacia oleracea L.).Theor Appl Genet. 2021 May;134(5):1319-1328. doi: 10.1007/s00122-021-03772-8. Epub 2021 Jan 29. Theor Appl Genet. 2021. PMID: 33515081
-
Comparative transcriptomic analyses of powdery mildew resistant and susceptible cultivated cucumber (Cucumis sativus L.) varieties to identify the genes involved in the resistance to Sphaerotheca fuliginea infection.PeerJ. 2020 Apr 17;8:e8250. doi: 10.7717/peerj.8250. eCollection 2020. PeerJ. 2020. PMID: 32337096 Free PMC article.
-
Two QTLs controlling Clubroot resistance identified from Bulked Segregant Sequencing in Pakchoi (Brassica campestris ssp. chinensis Makino).Sci Rep. 2019 Jun 25;9(1):9228. doi: 10.1038/s41598-019-44724-z. Sci Rep. 2019. PMID: 31239512 Free PMC article.
References
-
- Sitterly WR, Keinath AP. Gummy stem blight. Compend Cucurbit Dis. 1996;1996:27–8.
-
- Feng H, Shuju L. Advance and prospect in cucumber breeding research in China. Sci Agric Sin. 2000;33:100–2.
-
- Kooistra E. Powdery mildew resistance in cucumber. Euphytica. 1968;17:236–44.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

