Reexamining the Germination Phenotypes of Several Clostridium difficile Strains Suggests Another Role for the CspC Germinant Receptor
- PMID: 26668265
- PMCID: PMC4810609
- DOI: 10.1128/JB.00908-15
Reexamining the Germination Phenotypes of Several Clostridium difficile Strains Suggests Another Role for the CspC Germinant Receptor
Abstract
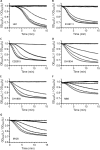
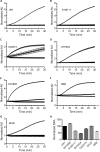
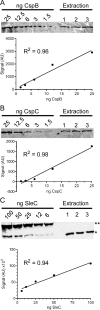
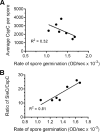
Clostridium difficile spore germination is essential for colonization and disease. The signals that initiate C. difficile spore germination are a combination of taurocholic acid (a bile acid) and glycine. Interestingly, the chenodeoxycholic acid class (CDCA) bile acids competitively inhibit taurocholic acid-mediated germination, suggesting that compounds that inhibit spore germination could be developed into drugs that prophylactically prevent C. difficile infection or reduce recurring disease. However, a recent report called into question the utility of such a strategy to prevent infection by describing C. difficile strains that germinated in the apparent absence of bile acids or germinated in the presence of the CDCA inhibitor. Because the mechanisms of C. difficile spore germination are beginning to be elucidated, the mechanism of germination in these particular strains could yield important information on how C. difficile spores initiate germination. Therefore, we quantified the interaction of these strains with taurocholic acid and CDCA, the rates of spore germination, the release of DPA from the spore core, and the abundance of the germinant receptor complex (CspC, CspB, and SleC). We found that strains previously observed to germinate in the absence of taurocholic acid correspond to more potent 50% effective concentrations (EC50 values; the concentrations that achieve a half-maximum germination rate) of the germinant and are still inhibited by CDCA, possibly explaining the previous observations. By comparing the germination kinetics and the abundance of proteins in the germinant receptor complex, we revised our original model for CspC-mediated activation of spore germination and propose that CspC may activate spore germination and then inhibit downstream processes.
Importance: Clostridium difficile forms metabolically dormant spores that persist in the health care environment. In susceptible hosts, C. difficile spores germinate in response to certain bile acids and glycine. Blocking germination by C. difficile spores is an attractive strategy to prevent the initiation of disease or to block recurring infection. However, certain C. difficile strains have been identified whose spores germinate in the absence of bile acids or are not blocked by known inhibitors of C. difficile spore germination (calling into question the utility of such strategies). Here, we further investigate these strains and reestablish that bile acid activators and inhibitors of germination affect these strains and use these data to suggest another role for the C. difficile bile acid germinant receptor.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Spore Cortex Hydrolysis Precedes Dipicolinic Acid Release during Clostridium difficile Spore Germination.J Bacteriol. 2015 Jul;197(14):2276-83. doi: 10.1128/JB.02575-14. Epub 2015 Apr 27. J Bacteriol. 2015. PMID: 25917906 Free PMC article.
-
Revisiting the Role of Csp Family Proteins in Regulating Clostridium difficile Spore Germination.J Bacteriol. 2017 Oct 17;199(22):e00266-17. doi: 10.1128/JB.00266-17. Print 2017 Nov 15. J Bacteriol. 2017. PMID: 28874406 Free PMC article.
-
The requirement for co-germinants during Clostridium difficile spore germination is influenced by mutations in yabG and cspA.PLoS Pathog. 2019 Apr 3;15(4):e1007681. doi: 10.1371/journal.ppat.1007681. eCollection 2019 Apr. PLoS Pathog. 2019. PMID: 30943268 Free PMC article.
-
A Revised Understanding of Clostridioides difficile Spore Germination.Trends Microbiol. 2020 Sep;28(9):744-752. doi: 10.1016/j.tim.2020.03.004. Epub 2020 Apr 23. Trends Microbiol. 2020. PMID: 32781028 Review.
-
Updates to Clostridium difficile Spore Germination.J Bacteriol. 2018 Jul 25;200(16):e00218-18. doi: 10.1128/JB.00218-18. Print 2018 Aug 15. J Bacteriol. 2018. PMID: 29760211 Free PMC article. Review.
Cited by
-
Impact of microbial derived secondary bile acids on colonization resistance against Clostridium difficile in the gastrointestinal tract.Anaerobe. 2016 Oct;41:44-50. doi: 10.1016/j.anaerobe.2016.05.003. Epub 2016 May 7. Anaerobe. 2016. PMID: 27163871 Free PMC article. Review.
-
Germinants and Their Receptors in Clostridia.J Bacteriol. 2016 Sep 22;198(20):2767-75. doi: 10.1128/JB.00405-16. Print 2016 Oct 15. J Bacteriol. 2016. PMID: 27432831 Free PMC article. Review.
-
Imaging Clostridioides difficile Spore Germination and Germination Proteins.J Bacteriol. 2022 Jul 19;204(7):e0021022. doi: 10.1128/jb.00210-22. Epub 2022 Jun 28. J Bacteriol. 2022. PMID: 35762766 Free PMC article.
-
Butyrate enhances Clostridioides difficile sporulation in vitro.J Bacteriol. 2023 Sep 26;205(9):e0013823. doi: 10.1128/jb.00138-23. Epub 2023 Sep 1. J Bacteriol. 2023. PMID: 37655912 Free PMC article.
-
Chemical and Stress Resistances of Clostridium difficile Spores and Vegetative Cells.Front Microbiol. 2016 Oct 26;7:1698. doi: 10.3389/fmicb.2016.01698. eCollection 2016. Front Microbiol. 2016. PMID: 27833595 Free PMC article.
References
-
- Jump RLP, Pultz MJ, Donskey CJ. 2007. Vegetative Clostridium difficile survives in room air on moist surfaces and in gastric contents with reduced acidity: a potential mechanism to explain the association between proton pump inhibitors and C. difficile-associated diarrhea? Antimicrob Agents Chemother 51:2883–2887. doi:10.1128/AAC.01443-06. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

