TspanC8 Tetraspanins and A Disintegrin and Metalloprotease 10 (ADAM10) Interact via Their Extracellular Regions: EVIDENCE FOR DISTINCT BINDING MECHANISMS FOR DIFFERENT TspanC8 PROTEINS
- PMID: 26668317
- PMCID: PMC4751363
- DOI: 10.1074/jbc.M115.703058
TspanC8 Tetraspanins and A Disintegrin and Metalloprotease 10 (ADAM10) Interact via Their Extracellular Regions: EVIDENCE FOR DISTINCT BINDING MECHANISMS FOR DIFFERENT TspanC8 PROTEINS
Abstract
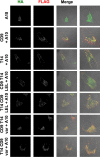
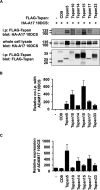
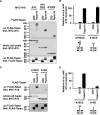
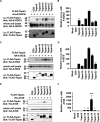
A disintegrin and metalloprotease 10 (ADAM10) is a ubiquitously expressed transmembrane metalloprotease that cleaves the extracellular regions from its transmembrane substrates. ADAM10 is essential for embryonic development and is implicated in cancer, Alzheimer, and inflammatory diseases. The tetraspanins are a superfamily of 33 four-transmembrane proteins in mammals, of which the TspanC8 subgroup (Tspan5, 10, 14, 15, 17, and 33) promote ADAM10 intracellular trafficking and enzymatic maturation. However, the interaction between TspanC8s and ADAM10 has only been demonstrated in overexpression systems and the interaction mechanism remains undefined. To address these issues, an antibody was developed to Tspan14, which was used to show co-immunoprecipitation of Tspan14 with ADAM10 in primary human cells. Chimeric Tspan14 constructs demonstrated that the large extracellular loop of Tspan14 mediated its co-immunoprecipitation with ADAM10, and promoted ADAM10 maturation and trafficking to the cell surface. Chimeric ADAM10 constructs showed that membrane-proximal stalk, cysteine-rich, and disintegrin domains of ADAM10 mediated its co-immunoprecipitation with Tspan14 and other TspanC8s. This TspanC8-interacting region was required for ADAM10 exit from the endoplasmic reticulum. Truncated ADAM10 constructs revealed differential TspanC8 binding requirements for the stalk, cysteine-rich, and disintegrin domains. Moreover, Tspan15 was the only TspanC8 to promote cleavage of the ADAM10 substrate N-cadherin, whereas Tspan14 was unique in reducing cleavage of the platelet collagen receptor GPVI. These findings suggest that ADAM10 may adopt distinct conformations in complex with different TspanC8s, which could impact on substrate selectivity. Furthermore, this study identifies regions of TspanC8s and ADAM10 for potential interaction-disrupting therapeutic targeting.
Keywords: ADAM; GPVI; N-cadherin; TspanC8; cell surface enzyme; endothelial cell; metalloprotease; platelet; shedding; tetraspanin.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures







References
-
- Saftig P., and Reiss K. (2011) The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17: Novel drug targets with therapeutic potential? Eur. J. Cell Biol. 90, 527–535 - PubMed
-
- Dreymueller D., Pruessmeyer J., Groth E., and Ludwig A. (2012) The role of ADAM-mediated shedding in vascular biology. Eur. J. Cell Biol. 91, 472–485 - PubMed
-
- Hartmann D., de Strooper B., Serneels L., Craessaerts K., Herreman A., Annaert W., Umans L., Lübke T., Lena Illert A., Figura, von K., and Saftig P. (2002) The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for α-secretase activity in fibroblasts. Hum. Mol. Genet. 11, 2615–2624 - PubMed
-
- Bender M., Hofmann S., Stegner D., Chalaris A., Bösl M., Braun A., Scheller J., Rose-John S., and Nieswandt B. (2010) Differentially regulated GPVI ectodomain shedding by multiple platelet-expressed proteinases. Blood 116, 3347–3355 - PubMed
-
- Gardiner E. E., Karunakaran D., Shen Y., Arthur J. F., Andrews R. K., and Berndt M. C. (2007) Controlled shedding of platelet glycoprotein (GP)VI and GPIb-IX-V by ADAM family metalloproteinases. J. Thromb. Haemost. 5, 1530–1537 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

