The Murine Lung Microbiome Changes During Lung Inflammation and Intranasal Vancomycin Treatment
- PMID: 26668669
- PMCID: PMC4676059
- DOI: 10.2174/1874285801509010167
The Murine Lung Microbiome Changes During Lung Inflammation and Intranasal Vancomycin Treatment
Abstract
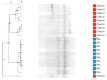
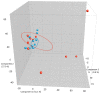
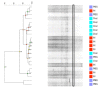
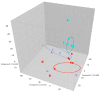
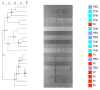
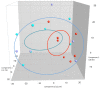
Most microbiome research related to airway diseases has focused on the gut microbiome. This is despite advances in culture independent microbial identification techniques revealing that even healthy lungs possess a unique dynamic microbiome. This conceptual change raises the question; if lung diseases could be causally linked to local dysbiosis of the local lung microbiota. Here, we manipulate the murine lung and gut microbiome, in order to show that the lung microbiota can be changed experimentally. We have used four different approaches: lung inflammation by exposure to carbon nano-tube particles, oral probiotics and oral or intranasal exposure to the antibiotic vancomycin. Bacterial DNA was extracted from broncho-alveolar and nasal lavage fluids, caecum samples and compared by DGGE. Our results show that: the lung microbiota is sex dependent and not just a reflection of the gut microbiota, and that induced inflammation can change lung microbiota. This change is not transferred to offspring. Oral probiotics in adult mice do not change lung microbiome detectible by DGGE. Nasal vancomycin can change the lung microbiome preferentially, while oral exposure does not. These observations should be considered in future studies of the causal relationship between lung microbiota and lung diseases.
Keywords: Antibiotic; Denaturing gradient gel electrophoresis; broncho-alveolar lavage; carbon nanotubes; probiotic.
Figures




References
-
- Huang Y.J., Nelson C.E., Brodie E.L., Desantis T.Z., Baek M.S., Liu J., Woyke T., Allgaier M., Bristow J., Wiener-Kronish J.P., Sutherland E.R., King T.S., Icitovic N., Martin R.J., Calhoun W.J., Castro M., Denlinger L.C., Dimango E., Kraft M., Peters S.P., Wasserman S.I., Wechsler M.E., Boushey H.A., Lynch S.V., National Heart, Lung, and Blood Institute’s Asthma Clinical Research Network Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J. Allergy Clin. Immunol. 2011;127(2):372–381.e1, 3. doi: 10.1016/j.jaci.2010.10.048. - DOI - PMC - PubMed
-
- Morris A., Beck J.M., Schloss P.D., Campbell T.B., Crothers K., Curtis J.L., Flores S.C., Fontenot A.P., Ghedin E., Huang L., Jablonski K., Kleerup E., Lynch S.V., Sodergren E., Twigg H., Young V.B., Bassis C.M., Venkataraman A., Schmidt T.M., Weinstock G.M., Lung HIV Microbiome Project Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 2013;187(10):1067–1075. doi: 10.1164/rccm.201210-1913OC. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
