A donor-specific epigenetic classifier for acute graft-versus-host disease severity in hematopoietic stem cell transplantation
- PMID: 26669438
- PMCID: PMC4681168
- DOI: 10.1186/s13073-015-0246-z
A donor-specific epigenetic classifier for acute graft-versus-host disease severity in hematopoietic stem cell transplantation
Abstract
Background: Allogeneic hematopoietic stem cell transplantation (HSCT) is a curative treatment for many hematological conditions. Acute graft-versus-host disease (aGVHD) is a prevalent immune-mediated complication following HSCT. Current diagnostic biomarkers that correlate with aGVHD severity, progression, and therapy response in graft recipients are insufficient. Here, we investigated whether epigenetic marks measured in peripheral blood of healthy graft donors stratify aGVHD severity in human leukocyte antigen (HLA)-matched sibling recipients prior to T cell-depleted HSCT.
Methods: We measured DNA methylation levels genome-wide at single-nucleotide resolution in peripheral blood of 85 HSCT donors, matched to recipients with various transplant outcomes, with Illumina Infinium HumanMethylation450 BeadChips.
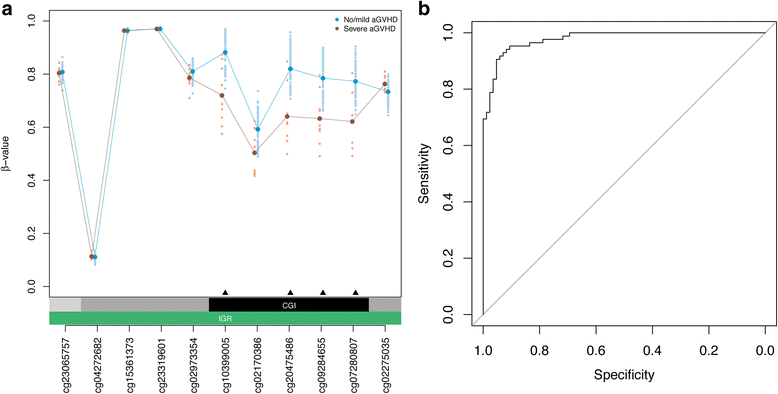
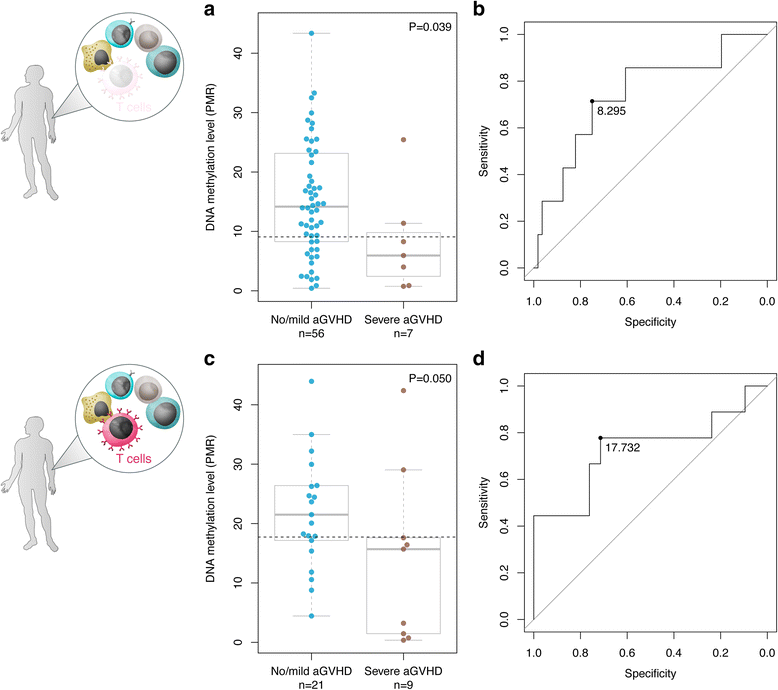
Results: Using genome-wide DNA methylation profiling, we showed that epigenetic signatures underlying aGVHD severity in recipients correspond to immune pathways relevant to aGVHD etiology. We discovered 31 DNA methylation marks in donors that associated with aGVHD severity status in recipients, and demonstrated strong predictive performance of these markers in internal cross-validation experiments (AUC = 0.98, 95% CI = 0.96-0.99). We replicated the top-ranked CpG classifier using an alternative, clinical DNA methylation assay (P = 0.039). In an independent cohort of 32 HSCT donors, we demonstrated the utility of the epigenetic classifier in the context of a T cell-replete conditioning regimen (P = 0.050).
Conclusions: Our findings suggest that epigenetic typing of HSCT donors in a clinical setting may be used in conjunction with HLA genotyping to inform both donor selection and transplantation strategy, with the ultimate aim of improving patient outcome.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

