An Automated Miniaturized Method to Perform and Analyze Antimicrobial Drug Synergy Assays
- PMID: 26669516
- PMCID: PMC4761817
- DOI: 10.1089/adt.2015.672
An Automated Miniaturized Method to Perform and Analyze Antimicrobial Drug Synergy Assays
Abstract
In the light of emerging antibiotic resistance mechanisms found in bacteria throughout the world, discovery of drugs that potentiate the effect of currently available antibiotics remains an important aspect of pharmaceutical research in the 21st century. Well-established clinical tests exist to determine synergy in vitro, but these are only optimal for low-throughput experimentation while leaving analysis of results and interpretation of high-throughput microscale assays poorly standardized. Here, we describe a miniaturized broth microdilution checkerboard assay and data analysis method in 384-well plate format that conforms to the Clinical Laboratory and Standards Institute (CLSI) methods. This method has been automated and developed to rapidly determine the synergism of current antibiotics with various beta-lactamase inhibitors emerging from our antimicrobial research efforts. This technique increases test throughput and integrity of results, and saves test compound and labor. We facilitated the interpretation of results with an automated analysis tool allowing us to rapidly qualify inter- and intraplate robustness, determine efficacy of multiple antibiotics at the same time, and standardize the results of synergy interpretation. This procedure should enhance high-throughput antimicrobial drug discovery and supersedes former techniques.
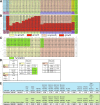
Figures


Similar articles
-
Miniaturized Checkerboard Assays to Measure Antibiotic Interactions.Methods Mol Biol. 2019;1939:3-9. doi: 10.1007/978-1-4939-9089-4_1. Methods Mol Biol. 2019. PMID: 30848453
-
Synergy of β-Lactams with Vancomycin against Methicillin-Resistant Staphylococcus aureus: Correlation of Disk Diffusion and Checkerboard Methods.J Clin Microbiol. 2016 Mar;54(3):565-8. doi: 10.1128/JCM.01779-15. Epub 2015 Dec 16. J Clin Microbiol. 2016. PMID: 26677253 Free PMC article.
-
Highly parallelized droplet cultivation and prioritization of antibiotic producers from natural microbial communities.Elife. 2021 Mar 25;10:e64774. doi: 10.7554/eLife.64774. Elife. 2021. PMID: 33764297 Free PMC article.
-
Correlation of the results of antibiotic synergy and susceptibility testing in vitro with results in experimental mouse infections.Crit Rev Microbiol. 1982;10(1):1-76. doi: 10.3109/10408418209113505. Crit Rev Microbiol. 1982. PMID: 6756787 Review.
-
[The implementation of the broth microdilution method to determine bacterial susceptibility to antimicrobial agents].Berl Munch Tierarztl Wochenschr. 2007 Jan-Feb;120(1-2):42-9. Berl Munch Tierarztl Wochenschr. 2007. PMID: 17290942 Review. German.
Cited by
-
Weakest-Link Dynamics Predict Apparent Antibiotic Interactions in a Model Cross-Feeding Community.Antimicrob Agents Chemother. 2020 Oct 20;64(11):e00465-20. doi: 10.1128/AAC.00465-20. Print 2020 Oct 20. Antimicrob Agents Chemother. 2020. PMID: 32778550 Free PMC article.
-
Assessment of tuberculosis drug efficacy using preclinical animal models and in vitro predictive techniques.NPJ Antimicrob Resist. 2024 Dec 16;2(1):49. doi: 10.1038/s44259-024-00066-z. NPJ Antimicrob Resist. 2024. PMID: 39843983 Free PMC article.
References
-
- Bush K: Antibacterial drug discovery in the 21st century. Clin Microbiol Infect. 2004;10 Suppl 4:10–17 - PubMed
-
- Blizzard TA, Chen H, Kim S, et al. : Side chain SAR of bicyclic beta-lactamase inhibitors (BLIs). 1. Discovery of a class C BLI for combination with imipinem. Bioorg Med Chem Lett 2010;20:918–921 - PubMed
-
- CDC: Antibiotic resistance threats in the United States, 2013. In: Services USDoHaH (ed.), pp. 1–114. CDC, Atlanta, Georgia, 2013
-
- European Centre for Disease Prevention and Control: Summary of the Latest Data on Antibiotic Resistance in the European Union, pp. 1–10, Stockholm, 2013
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

