Utilization of Benchtop Next Generation Sequencing Platforms Ion Torrent PGM and MiSeq in Noninvasive Prenatal Testing for Chromosome 21 Trisomy and Testing of Impact of In Silico and Physical Size Selection on Its Analytical Performance
- PMID: 26669558
- PMCID: PMC4692262
- DOI: 10.1371/journal.pone.0144811
Utilization of Benchtop Next Generation Sequencing Platforms Ion Torrent PGM and MiSeq in Noninvasive Prenatal Testing for Chromosome 21 Trisomy and Testing of Impact of In Silico and Physical Size Selection on Its Analytical Performance
Abstract
Objectives: The aims of this study were to test the utility of benchtop NGS platforms for NIPT for trisomy 21 using previously published z score calculation methods and to optimize the sample preparation and data analysis with use of in silico and physical size selection methods.
Methods: Samples from 130 pregnant women were analyzed by whole genome sequencing on benchtop NGS systems Ion Torrent PGM and MiSeq. The targeted yield of 3 million raw reads on each platform was used for z score calculation. The impact of in silico and physical size selection on analytical performance of the test was studied.
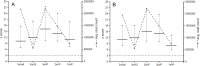
Results: Using a z score value of 3 as the cut-off, 98.11%-100% (104-106/106) specificity and 100% (24/24) sensitivity and 99.06%-100% (105-106/106) specificity and 100% (24/24) sensitivity were observed for Ion Torrent PGM and MiSeq, respectively. After in silico based size selection both platforms reached 100% specificity and sensitivity. Following the physical size selection z scores of tested trisomic samples increased significantly--p = 0.0141 and p = 0.025 for Ion Torrent PGM and MiSeq, respectively.
Conclusions: Noninvasive prenatal testing for chromosome 21 trisomy with the utilization of benchtop NGS systems led to results equivalent to previously published studies performed on high-to-ultrahigh throughput NGS systems. The in silico size selection led to higher specificity of the test. Physical size selection performed on isolated DNA led to significant increase in z scores. The observed results could represent a basis for increasing of cost effectiveness of the test and thus help with its penetration worldwide.
Conflict of interest statement
Figures



References
-
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350: 485–7. - PubMed
-
- Lo YM, Hjelm NM, Fidler C, Sargent IL, Murphy MF, Chamberlain PF, et al. Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. N Engl J Med. 1998;339: 1734–8. - PubMed
-
- Palomaki GE, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, Ehrich M, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med. 2011;13: 913–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

