Mutation Screening of 1,237 Cancer Genes across Six Model Cell Lines of Basal-Like Breast Cancer
- PMID: 26670335
- PMCID: PMC4684399
- DOI: 10.1371/journal.pone.0144528
Mutation Screening of 1,237 Cancer Genes across Six Model Cell Lines of Basal-Like Breast Cancer
Abstract
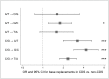
Basal-like breast cancer is an aggressive subtype generally characterized as poor prognosis and lacking the expression of the three most important clinical biomarkers, estrogen receptor, progesterone receptor, and HER2. Cell lines serve as useful model systems to study cancer biology in vitro and in vivo. We performed mutational profiling of six basal-like breast cancer cell lines (HCC38, HCC1143, HCC1187, HCC1395, HCC1954, and HCC1937) and their matched normal lymphocyte DNA using targeted capture and next-generation sequencing of 1,237 cancer-associated genes, including all exons, UTRs and upstream flanking regions. In total, 658 somatic variants were identified, of which 378 were non-silent (average 63 per cell line, range 37-146) and 315 were novel (not present in the Catalogue of Somatic Mutations in Cancer database; COSMIC). 125 novel mutations were confirmed by Sanger sequencing (59 exonic, 48 3'UTR and 10 5'UTR, 1 splicing), with a validation rate of 94% of high confidence variants. Of 36 mutations previously reported for these cell lines but not detected in our exome data, 36% could not be detected by Sanger sequencing. The base replacements C/G>A/T, C/G>G/C, C/G>T/A and A/T>G/C were significantly more frequent in the coding regions compared to the non-coding regions (OR 3.2, 95% CI 2.0-5.3, P<0.0001; OR 4.3, 95% CI 2.9-6.6, P<0.0001; OR 2.4, 95% CI 1.8-3.1, P<0.0001; OR 1.8, 95% CI 1.2-2.7, P = 0.024, respectively). The single nucleotide variants within the context of T[C]T/A[G]A and T[C]A/T[G]A were more frequent in the coding than in the non-coding regions (OR 3.7, 95% CI 2.2-6.1, P<0.0001; OR 3.8, 95% CI 2.0-7.2, P = 0.001, respectively). Copy number estimations were derived from the targeted regions and correlated well to Affymetrix SNP array copy number data (Pearson correlation 0.82 to 0.96 for all compared cell lines; P<0.0001). These mutation calls across 1,237 cancer-associated genes and identification of novel variants will aid in the design and interpretation of biological experiments using these six basal-like breast cancer cell lines.
Conflict of interest statement
Figures




References
-
- Ferlay J, Soerjomataram II, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. International journal of cancer Journal international du cancer. 2014. - PubMed
-
- Futreal PA, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, et al. BRCA1 mutations in primary breast and ovarian carcinomas. Science. 1994;266(5182):120–2. - PubMed
-
- Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science. 1994;265(5181):2088–90. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

