Impact of Anaerobiosis on Expression of the Iron-Responsive Fur and RyhB Regulons
- PMID: 26670385
- PMCID: PMC4676285
- DOI: 10.1128/mBio.01947-15
Impact of Anaerobiosis on Expression of the Iron-Responsive Fur and RyhB Regulons
Abstract
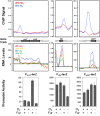
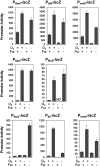
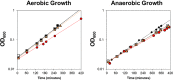
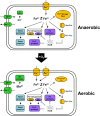
Iron, a major protein cofactor, is essential for most organisms. Despite the well-known effects of O2 on the oxidation state and solubility of iron, the impact of O2 on cellular iron homeostasis is not well understood. Here we report that in Escherichia coli K-12, the lack of O2 dramatically changes expression of genes controlled by the global regulators of iron homeostasis, the transcription factor Fur and the small RNA RyhB. Using chromatin immunoprecipitation sequencing (ChIP-seq), we found anaerobic conditions promote Fur binding to more locations across the genome. However, by expression profiling, we discovered that the major effect of anaerobiosis was to increase the magnitude of Fur regulation, leading to increased expression of iron storage proteins and decreased expression of most iron uptake pathways and several Mn-binding proteins. This change in the pattern of gene expression also correlated with an unanticipated decrease in Mn in anaerobic cells. Changes in the genes posttranscriptionally regulated by RyhB under aerobic and anaerobic conditions could be attributed to O2-dependent changes in transcription of the target genes: aerobic RyhB targets were enriched in iron-containing proteins associated with aerobic energy metabolism, whereas anaerobic RyhB targets were enriched in iron-containing anaerobic respiratory functions. Overall, these studies showed that anaerobiosis has a larger impact on iron homeostasis than previously anticipated, both by expanding the number of direct Fur target genes and the magnitude of their regulation and by altering the expression of genes predicted to be posttranscriptionally regulated by the small RNA RyhB under iron-limiting conditions.
Importance: Microbes and host cells engage in an "arms race" for iron, an essential nutrient that is often scarce in the environment. Studies of iron homeostasis have been key to understanding the control of iron acquisition and the downstream pathways that enable microbes to compete for this valuable resource. Here we report that O2 availability affects the gene expression programs of two Escherichia coli master regulators that function in iron homeostasis: the transcription factor Fur and the small RNA regulator RyhB. Fur appeared to be more active under anaerobic conditions, suggesting a change in the set point for iron homeostasis. RyhB preferentially targeted iron-containing proteins of respiration-linked pathways, which are differentially expressed under aerobic and anaerobic conditions. Such findings may be relevant to the success of bacteria within their hosts since zones of reduced O2 may actually reduce bacterial iron demands, making it easier to win the arms race for iron.
Copyright © 2015 Beauchene et al.
Figures



References
-
- Andrews S, Norton I, Salunkhe AS, Goodluck H, Aly WS, Mourad-Agha H, Cornelis P. 2013. Control of iron metabolism in bacteria. Met Ions Life Sci 12:203–239. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

