Starvation Induces Proteasome Autophagy with Different Pathways for Core and Regulatory Particles
- PMID: 26670610
- PMCID: PMC4751371
- DOI: 10.1074/jbc.M115.699124
Starvation Induces Proteasome Autophagy with Different Pathways for Core and Regulatory Particles
Abstract
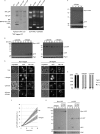
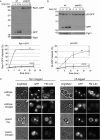
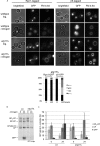
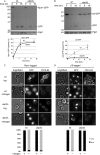
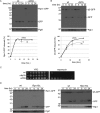
The proteasome is responsible for the degradation of many cellular proteins. If and how this abundant and normally stable complex is degraded by cells is largely unknown. Here we show that in yeast, upon nitrogen starvation, proteasomes are targeted for vacuolar degradation through autophagy. Using GFP-tagged proteasome subunits, we observed that autophagy of a core particle (CP) subunit depends on the deubiquitinating enzyme Ubp3, although a regulatory particle (RP) subunit does not. Furthermore, upon blocking of autophagy, RP remained largely nuclear, although CP largely localized to the cytosol as well as granular structures within the cytosol. In all, our data reveal a regulated process for the removal of proteasomes upon nitrogen starvation. This process involves CP and RP dissociation, nuclear export, and independent vacuolar targeting of CP and RP. Thus, in addition to the well characterized transcriptional up-regulation of genes encoding proteasome subunits, cells are also capable of down-regulating cellular levels of proteasomes through proteaphagy.
Keywords: autophagy; deubiquitylation (deubiquitination); nitrogen starvation; proteaphagy; proteasome; protein translocation; ribophagy; vacuole; yeast.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Warner J. R. (1999) The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

