Genome-wide DNA methylation profiling of peripheral blood mononuclear cells in irritable bowel syndrome
- PMID: 26670691
- PMCID: PMC4760882
- DOI: 10.1111/nmo.12741
Genome-wide DNA methylation profiling of peripheral blood mononuclear cells in irritable bowel syndrome
Abstract
Background: Irritable bowel syndrome (IBS) is a stress-sensitive disorder. Environmental factors including stress can trigger epigenetic changes, which have not been well-studied in IBS. We performed a pilot study investigating genome-wide DNA methylation of IBS patients and healthy controls (HCs) to identify potential epigenetic markers and associated pathways. Additionally, we investigated relationships of epigenetic changes in selected genes with clinical traits.
Methods: Twenty-seven IBS patients (59% women; 10 IBS-diarrhea, 8 IBS-constipation, 9 IBS-mixed) and 23 age- and sex-matched HCs were examined. DNA methylation from peripheral blood mononuclear cells (PBMCs) was measured using HM450 BeadChip, and representative methylation differences were confirmed by bisulphite sequencing. Gene expression was measured using quantitative PCR. Gastrointestinal (GI) and non-GI symptoms were measured using validated questionnaires. Associations were tested using non-parametric methods.
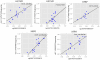
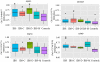
Key results: Genome-wide DNA methylation profiling of IBS patients compared with HCs identified 133 differentially methylated positions (DMPs) (mean difference ≥10%; p < 0.05). These genes were associated with gene ontology terms including glutathione metabolism related to oxidative stress and neuropeptide hormone activity. Validation by sequencing confirmed differential methylation of subcommissural organ (SCO)-Spondin (SSPO), glutathione-S-transferases mu 5 (GSTM5), and tubulin polymerization promoting protein genes. Methylation of two promoter CpGs in GSTM5 was associated with epigenetic silencing. Epigenetic changes in SSPO gene were positively correlated with hospital anxiety and depression scores in IBS patients (r > 0.4 and false discovery rate <0.05).
Conclusions & inferences: This study is the first to comprehensively explore the methylome of IBS patients. We identified DMPs in novel candidate genes which could provide new insights into disease mechanisms; however, these preliminary findings warrant confirmation in larger, independent studies.
Keywords: DNA methylation; bisulphite sequencing; human methylation 450 array; irritable bowel syndrome; oxidative stress.
© 2015 John Wiley & Sons Ltd.
Figures
References
-
- Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC. Functional bowel disorders. Gastroenterology. 2006;130:1480–91. doi:10.1053/j.gastro.2005.11.061. - PubMed
-
- Lovell RM, Ford AC. Global prevalence of and risk factors for irritable bowel syndrome: a meta-analysis. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. 2012;10:712–21. e4. doi:10.1016/j.cgh.2012.02.029. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials