BMDExpress Data Viewer - a visualization tool to analyze BMDExpress datasets
- PMID: 26671443
- PMCID: PMC5064610
- DOI: 10.1002/jat.3265
BMDExpress Data Viewer - a visualization tool to analyze BMDExpress datasets
Abstract
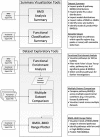
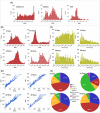
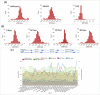
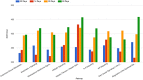
Regulatory agencies increasingly apply benchmark dose (BMD) modeling to determine points of departure for risk assessment. BMDExpress applies BMD modeling to transcriptomic datasets to identify transcriptional BMDs. However, graphing and analytical capabilities within BMDExpress are limited, and the analysis of output files is challenging. We developed a web-based application, BMDExpress Data Viewer (http://apps.sciome.com:8082/BMDX_Viewer/), for visualizing and graphing BMDExpress output files. The application consists of "Summary Visualization" and "Dataset Exploratory" tools. Through analysis of transcriptomic datasets of the toxicants furan and 4,4'-methylenebis(N,N-dimethyl)benzenamine, we demonstrate that the "Summary Visualization Tools" can be used to examine distributions of gene and pathway BMD values, and to derive a potential point of departure value based on summary statistics. By applying filters on enrichment P-values and minimum number of significant genes, the "Functional Enrichment Analysis" tool enables the user to select biological processes or pathways that are selectively perturbed by chemical exposure and identify the related BMD. The "Multiple Dataset Comparison" tool enables comparison of gene and pathway BMD values across multiple experiments (e.g., across timepoints or tissues). The "BMDL-BMD Range Plotter" tool facilitates the observation of BMD trends across biological processes or pathways. Through our case studies, we demonstrate that BMDExpress Data Viewer is a useful tool to visualize, explore and analyze BMDExpress output files. Visualizing the data in this manner enables rapid assessment of data quality, model fit, doses of peak activity, most sensitive pathway perturbations and other metrics that will be useful in applying toxicogenomics in risk assessment. © 2015 Her Majesty the Queen in Right of Canada. Journal of Applied Toxicology published by John Wiley & Sons, Ltd.
Keywords: 4,4′-methylenebis(N,N-dimethyl)benzenamine (MDMB); BMDExpress; benchmark dose (BMD); bioinformatics; data visualization; dose-response; furan; gene expression; genomics; human health risk assessment; microarray; transcriptomics.
© 2015 Her Majesty the Queen in Right of Canada. Journal of Applied Toxicology published by John Wiley & Sons, Ltd.
Figures


References
-
- Bertie AJ. 2002. Java applications for teaching statistics. MSOR Connect 2: ui–81.
-
- Bhat VS, Hester SD, Nesnow S, Eastmond DA. 2013. Concordance of transcriptional and apical benchmark dose levels for conazole‐induced liver effects in mice. Toxicol. Sci. 136: 205–215. - PubMed
-
- Black MB, Parks BB, Pluta L, Chu TM, Allen BC, Wolfinger RD, Thomas RS. 2014. Comparison of microarrays and RNA‐seq for gene expression analyses of dose‐response experiments. Toxicol. Sci. 137: 385–403. - PubMed
-
- Budtz‐Jorgensen E, Bellinger D, Lanphear B, Grandjean P, International Pooled Lead Study Investigators. 2013. An international pooled analysis for obtaining a benchmark dose for environmental lead exposure in children. Risk Anal. 33: 450–461. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

