The Time Scale of Recombination Rate Evolution in Great Apes
- PMID: 26671457
- PMCID: PMC5870646
- DOI: 10.1093/molbev/msv331
The Time Scale of Recombination Rate Evolution in Great Apes
Abstract
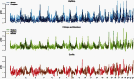
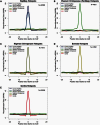
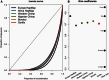
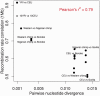
We present three linkage-disequilibrium (LD)-based recombination maps generated using whole-genome sequence data from 10 Nigerian chimpanzees, 13 bonobos, and 15 western gorillas, collected as part of the Great Ape Genome Project (Prado-Martinez J, et al. 2013. Great ape genetic diversity and population history. Nature 499:471-475). We also identified species-specific recombination hotspots in each group using a modified LDhot framework, which greatly improves statistical power to detect hotspots at varying strengths. We show that fewer hotspots are shared among chimpanzee subspecies than within human populations, further narrowing the time scale of complete hotspot turnover. Further, using species-specific PRDM9 sequences to predict potential binding sites (PBS), we show higher predicted PRDM9 binding in recombination hotspots as compared to matched cold spot regions in multiple great ape species, including at least one chimpanzee subspecies. We found that correlations between broad-scale recombination rates decline more rapidly than nucleotide divergence between species. We also compared the skew of recombination rates at centromeres and telomeres between species and show a skew from chromosome means extending as far as 10-15 Mb from chromosome ends. Further, we examined broad-scale recombination rate changes near a translocation in gorillas and found minimal differences as compared to other great ape species perhaps because the coordinates relative to the chromosome ends were unaffected. Finally, on the basis of multiple linear regression analysis, we found that various correlates of recombination rate persist throughout the African great apes including repeats, diversity, and divergence. Our study is the first to analyze within- and between-species genome-wide recombination rate variation in several close relatives.
Keywords: PRDM9; hotspots; primates; recombination.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

