Stochasticity and homeostasis in the E. coli replication and division cycle
- PMID: 26671779
- PMCID: PMC4680914
- DOI: 10.1038/srep18261
Stochasticity and homeostasis in the E. coli replication and division cycle
Abstract
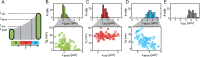
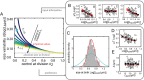
How cells correct for stochasticity to coordinate the chromosome replication and cellular division cycle is poorly understood. We used time-lapse microscopy and fluorescently labelled SeqA to determine the timing of birth, initiation, termination, and division, as well as cell size throughout the cell cycle. We found that the time between birth and initiation (B-period) compensates for stochastic variability in birth size and growth rate. The time between termination and division (D-period) also compensates for size and growth variability, invalidating the notion that replication initiation is the principal trigger for cell division. In contrast, the time between initiation and termination (C-period) did not display such compensations. Interestingly, the C-period did show small but systematic decreases for cells that spontaneously grew faster, which suggests a coupling between metabolic fluctuations and replication. An auto-regressive theoretical framework was employed to compare different possible models of sub-period control.
Figures


References
-
- Michelsen O., de Mattos M. J. T., Jensen P. R. & Hansen F. G. Precise determinations of C and D periods by flow cytometry in Escherichia coli K-12 and B/r. Microbiol-Sgm 149, 1001–1010, (2003). - PubMed
-
- Koch A. L. & Schaechter M. A model for statistics of the cell division process. J Gen Microbiol 29, 435–454, (1962). - PubMed

