Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences
- PMID: 26673039
- PMCID: PMC4864062
- DOI: 10.2217/epi.15.114
Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences
Abstract
Aim: DNA methylation is the best known epigenetic mark. Cancer and other pathologies show an altered DNA methylome. However, delivering complete DNA methylation maps is compromised by the price and labor-intensive interpretation of single nucleotide methods.
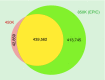
Material & methods: Following the success of the HumanMethylation450 BeadChip (Infinium) methylation microarray (450K), we report the technical and biological validation of the newly developed MethylationEPIC BeadChip (Infinium) microarray that covers over 850,000 CpG methylation sites (850K). The 850K microarray contains >90% of the 450K sites, but adds 333,265 CpGs located in enhancer regions identified by the ENCODE and FANTOM5 projects.
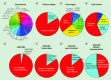
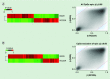
Results & conclusion: The 850K array demonstrates high reproducibility at the 450K CpG sites, is consistent among technical replicates, is reliable in the matched study of fresh frozen versus formalin-fixed paraffin-embeded samples and is also useful for 5-hydroxymethylcytosine. These results highlight the value of the MethylationEPIC BeadChip as a useful tool for the analysis of the DNA methylation profile of the human genome.
Keywords: 5-hydroxymethylcytosine; CpG sites; DNA methylation; epigenetics; microarray; paraffin; validation.
Conflict of interest statement
Financial & competing interests disclosure This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement No 640696) and under the European Community's Seventh Framework Programme (FP7/2007–2013)/ERC grant agreement no. 268626 and from Ministerio de Economía y Competitividad (MINECO), co-financed by the European Development Regional Fund, ‘A way to achieve Europe’ ERDF, under grant no. SAF2014-55000-R, the Instituto de Salud Carlos III (ISCIII) by the Spanish Cancer Research Network (RTICC) no. RD12/0036/0039, the Cellex Foundation, the AGAUR 2014SGR633 grant and the Health and Science Departments of the Catalan government (Generalitat de Catalunya). ME is an ICREA Research Professor. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Figures


References
-
- Feinberg AP. An epigenetic approach to cancer etiology. Cancer J. 2007;13(1):70–74. - PubMed
-
- Weidman JR, Dolinoy DC, Murphy SK, Jirtle RL. Cancer susceptibility: epigenetic manifestation of environmental exposures. Cancer J. 2007;13(1):9–16. - PubMed
-
- Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358(11):1148–1159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
