The Msi Family of RNA-Binding Proteins Function Redundantly as Intestinal Oncoproteins
- PMID: 26673327
- PMCID: PMC4894540
- DOI: 10.1016/j.celrep.2015.11.022
The Msi Family of RNA-Binding Proteins Function Redundantly as Intestinal Oncoproteins
Abstract
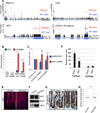
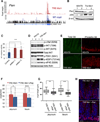
Members of the Msi family of RNA-binding proteins have recently emerged as potent oncoproteins in a range of malignancies. MSI2 is highly expressed in hematopoietic cancers, where it is required for disease maintenance. In contrast to the hematopoietic system, colorectal cancers can express both Msi family members, MSI1 and MSI2. Here, we demonstrate that, in the intestinal epithelium, Msi1 and Msi2 have analogous oncogenic effects. Further, comparison of Msi1/2-induced gene expression programs and transcriptome-wide analyses of Msi1/2-RNA-binding targets reveal significant functional overlap, including induction of the PDK-Akt-mTORC1 axis. Ultimately, we demonstrate that concomitant loss of function of both MSI family members is sufficient to abrogate the growth of human colorectal cancer cells, and Msi gene deletion inhibits tumorigenesis in several mouse models of intestinal cancer. Our findings demonstrate that MSI1 and MSI2 act as functionally redundant oncoproteins required for the ontogeny of intestinal cancers.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. - PubMed
-
- Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21WAF-1. Mol Cell Neurosci. 2006;31:85–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

