Transcriptional, post-transcriptional and chromatin-associated regulation of pri-miRNAs, pre-miRNAs and moRNAs
- PMID: 26673698
- PMCID: PMC4838339
- DOI: 10.1093/nar/gkv1354
Transcriptional, post-transcriptional and chromatin-associated regulation of pri-miRNAs, pre-miRNAs and moRNAs
Abstract
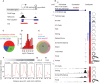
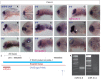
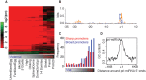
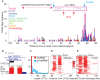
MicroRNAs (miRNAs) play a major role in the post-transcriptional regulation of target genes, especially in development and differentiation. Our understanding about the transcriptional regulation of miRNA genes is limited by inadequate annotation of primary miRNA (pri-miRNA) transcripts. Here, we used CAGE-seq and RNA-seq to provide genome-wide identification of the pri-miRNA core promoter repertoire and its dynamic usage during zebrafish embryogenesis. We assigned pri-miRNA promoters to 152 precursor-miRNAs (pre-miRNAs), the majority of which were supported by promoter associated post-translational histone modifications (H3K4me3, H2A.Z) and RNA polymerase II (RNAPII) occupancy. We validated seven miR-9 pri-miRNAs by in situ hybridization and showed similar expression patterns as mature miR-9. In addition, processing of an alternative intronic promoter of miR-9-5 was validated by 5' RACE PCR. Developmental profiling revealed a subset of pri-miRNAs that are maternally inherited. Moreover, we show that promoter-associated H3K4me3, H2A.Z and RNAPII marks are not only present at pri-miRNA promoters but are also specifically enriched at pre-miRNAs, suggesting chromatin level regulation of pre-miRNAs. Furthermore, we demonstrated that CAGE-seq also detects 3'-end processing of pre-miRNAs on Drosha cleavage site that correlates with miRNA-offset RNAs (moRNAs) production and provides a new tool for detecting Drosha processing events and predicting pre-miRNA processing by a genome-wide assay.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. - PubMed
-
- He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. - PubMed
-
- Borchert G.M., Lanier W., Davidson B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

