A transcription-independent epigenetic mechanism is associated with antigenic switching in Trypanosoma brucei
- PMID: 26673706
- PMCID: PMC4838347
- DOI: 10.1093/nar/gkv1459
A transcription-independent epigenetic mechanism is associated with antigenic switching in Trypanosoma brucei
Abstract
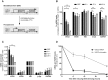
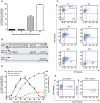
Antigenic variation in Trypanosoma brucei relies on periodic switching of variant surface glycoproteins (VSGs), which are transcribed monoallelically by RNA polymerase I from one of about 15 bloodstream expression sites (BES). Chromatin of the actively transcribed BES is depleted of nucleosomes, but it is unclear if this open conformation is a mere consequence of a high rate of transcription, or whether it is maintained by a transcription-independent mechanism. Using an inducible BES-silencing reporter strain, we observed that chromatin of the active BES remains open for at least 24 hours after blocking transcription. This conformation is independent of the cell-cycle stage, but dependent upon TDP1, a high mobility group box protein. For two days after BES silencing, we detected a transient and reversible derepression of several silent BESs within the population, suggesting that cells probe other BESs before commitment to one, which is complete by 48 hours. FACS sorting and subsequent subcloning confirmed that probing cells are switching intermediates capable of returning to the original BES, switch to the probed BES or to a different BES. We propose that regulation of BES chromatin structure is an epigenetic mechanism important for successful antigenic switching.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures






References
-
- Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. - PubMed
-
- McStay B., Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008;24:131–157. - PubMed
-
- Wittner M., Hamperl S., Stockl U., Seufert W., Tschochner H., Milkereit P., Griesenbeck J. Establishment and maintenance of alternative chromatin states at a multicopy gene locus. Cell. 2011;145:543–554. - PubMed
-
- Figueiredo L.M., Cross G.A., Janzen C.J. Epigenetic regulation in African trypanosomes: a new kid on the block. Nat. Rev. Microbiol. 2009;7:504–513. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

