canSAR: an updated cancer research and drug discovery knowledgebase
- PMID: 26673713
- PMCID: PMC4702774
- DOI: 10.1093/nar/gkv1030
canSAR: an updated cancer research and drug discovery knowledgebase
Abstract
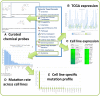
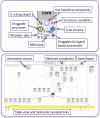
canSAR (http://cansar.icr.ac.uk) is a publicly available, multidisciplinary, cancer-focused knowledgebase developed to support cancer translational research and drug discovery. canSAR integrates genomic, protein, pharmacological, drug and chemical data with structural biology, protein networks and druggability data. canSAR is widely used to rapidly access information and help interpret experimental data in a translational and drug discovery context. Here we describe major enhancements to canSAR including new data, improved search and browsing capabilities, new disease and cancer cell line summaries and new and enhanced batch analysis tools.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
canSAR: updated cancer research and drug discovery knowledgebase.Nucleic Acids Res. 2014 Jan;42(Database issue):D1040-7. doi: 10.1093/nar/gkt1182. Epub 2013 Dec 3. Nucleic Acids Res. 2014. PMID: 24304894 Free PMC article.
-
canSAR: update to the cancer translational research and drug discovery knowledgebase.Nucleic Acids Res. 2019 Jan 8;47(D1):D917-D922. doi: 10.1093/nar/gky1129. Nucleic Acids Res. 2019. PMID: 30496479 Free PMC article.
-
canSAR: update to the cancer translational research and drug discovery knowledgebase.Nucleic Acids Res. 2021 Jan 8;49(D1):D1074-D1082. doi: 10.1093/nar/gkaa1059. Nucleic Acids Res. 2021. PMID: 33219674 Free PMC article.
-
Reverse Translational Research of ABCG2 (BCRP) in Human Disease and Drug Response.Clin Pharmacol Ther. 2018 Feb;103(2):233-242. doi: 10.1002/cpt.903. Epub 2017 Nov 28. Clin Pharmacol Ther. 2018. PMID: 29023674 Free PMC article. Review.
-
Targeting protein-protein interactions as an anticancer strategy.Trends Pharmacol Sci. 2013 Jul;34(7):393-400. doi: 10.1016/j.tips.2013.04.007. Epub 2013 May 29. Trends Pharmacol Sci. 2013. PMID: 23725674 Free PMC article. Review.
Cited by
-
Free Accessible Databases as a Source of Information about Food Components and Other Compounds with Anticancer Activity⁻Brief Review.Molecules. 2019 Feb 22;24(4):789. doi: 10.3390/molecules24040789. Molecules. 2019. PMID: 30813234 Free PMC article. Review.
-
Objective, Quantitative, Data-Driven Assessment of Chemical Probes.Cell Chem Biol. 2018 Feb 15;25(2):194-205.e5. doi: 10.1016/j.chembiol.2017.11.004. Epub 2017 Dec 14. Cell Chem Biol. 2018. PMID: 29249694 Free PMC article.
-
Leveraging Human Genetics to Guide Cancer Drug Development.JCO Clin Cancer Inform. 2018 Dec;2:1-11. doi: 10.1200/CCI.18.00077. JCO Clin Cancer Inform. 2018. PMID: 30652614 Free PMC article.
-
Molecular Signatures of Fusion Proteins in Cancer.ACS Pharmacol Transl Sci. 2019 Mar 20;2(2):122-133. doi: 10.1021/acsptsci.9b00019. eCollection 2019 Apr 12. ACS Pharmacol Transl Sci. 2019. PMID: 32219217 Free PMC article.
-
Analysis of impact metrics for the Protein Data Bank.Sci Data. 2018 Oct 16;5:180212. doi: 10.1038/sdata.2018.212. Sci Data. 2018. PMID: 30325351 Free PMC article.
References
-
- Patel M.N., Halling-Brown M.D., Tym J.E., Workman P., Al-Lazikani B. Objective assessment of cancer genes for drug discovery. Nat. Rev. Drug Discov. 2013;12:35–50. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

