Large-scale targeted sequencing comparison highlights extreme genetic heterogeneity in nephronophthisis-related ciliopathies
- PMID: 26673778
- PMCID: PMC5057575
- DOI: 10.1136/jmedgenet-2015-103304
Large-scale targeted sequencing comparison highlights extreme genetic heterogeneity in nephronophthisis-related ciliopathies
Abstract
Background: The term nephronophthisis-related ciliopathies (NPHP-RC) describes a group of rare autosomal-recessive cystic kidney diseases, characterised by broad genetic and clinical heterogeneity. NPHP-RC is frequently associated with extrarenal manifestations and accounts for the majority of genetically caused chronic kidney disease (CKD) during childhood and adolescence. Generation of a molecular diagnosis has been impaired by this broad genetic heterogeneity. However, recently developed high-throughput exon sequencing techniques represent powerful and efficient tools to screen large cohorts for dozens of causative genes.
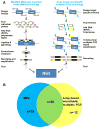
Methods: Therefore, we performed massively multiplexed targeted sequencing using the modified molecular inversion probe strategy (MIPs) in an international cohort of 384 patients diagnosed with NPHP-RC.
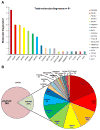
Results: As a result, we established the molecular diagnoses in 81/384 unrelated individuals (21.1%). We detected 127 likely disease-causing mutations in 18 of 34 evaluated NPHP-RC genes, 22 of which were novel. We further compared a subgroup of current findings to the results of a previous study in which we used an array-based microfluidic PCR technology in the same cohort. While 78 likely disease-causing mutations were previously detected by the array-based microfluidic PCR, the MIPs approach identified 94 likely pathogenic mutations. Compared with the previous approach, MIPs redetected 66 out of 78 variants and 28 previously unidentified variants, for a total of 94 variants.
Conclusions: In summary, we demonstrate that the modified MIPs technology is a useful approach to screen large cohorts for a multitude of established NPHP genes in order to identify the underlying molecular cause. Combined application of two independent library preparation and sequencing techniques, however, may still be indicated for Mendelian diseases with extensive genetic heterogeneity in order to further increase diagnostic sensitivity.
Keywords: Diagnostics; Genetics; Molecular genetics; Renal Medicine.
Published by the BMJ Publishing Group Limited. For permission to use (where not already granted under a licence) please go to http://www.bmj.com/company/products-services/rights-and-licensing/
Conflict of interest statement
output.txt These authors contributed equally to this work
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
