Protein aggregation, structural disorder and RNA-binding ability: a new approach for physico-chemical and gene ontology classification of multiple datasets
- PMID: 26673865
- PMCID: PMC4681139
- DOI: 10.1186/s12864-015-2280-z
Protein aggregation, structural disorder and RNA-binding ability: a new approach for physico-chemical and gene ontology classification of multiple datasets
Abstract
Background: Comparison between multiple protein datasets requires the choice of an appropriate reference system and a number of variables to describe their differences. Here we introduce an innovative approach to discriminate multiple protein datasets (multiCM) and to measure enrichments in gene ontology terms (cleverGO) using semantic similarities.
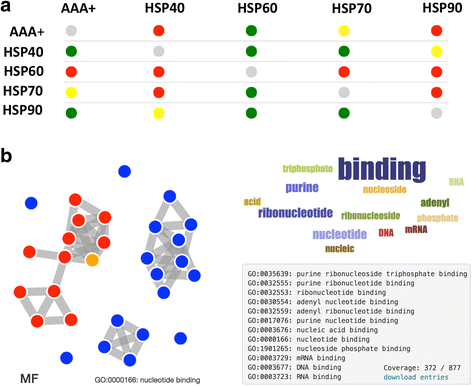
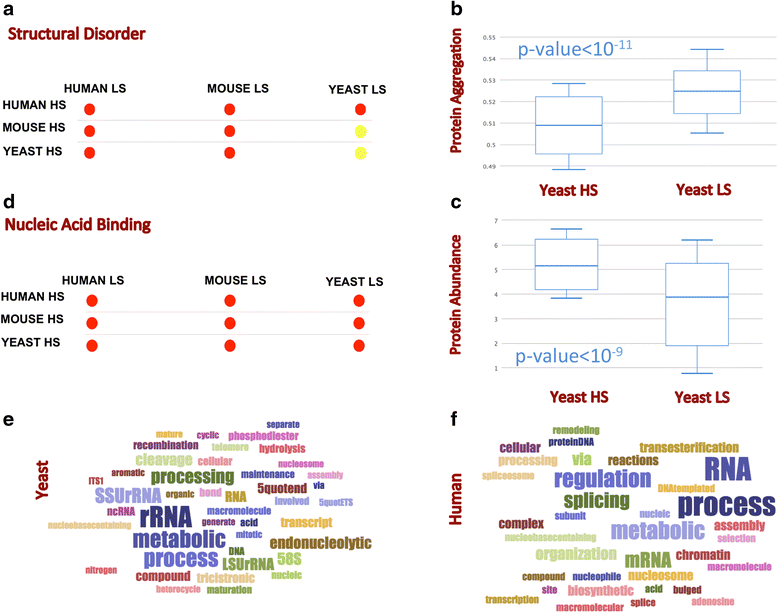
Results: We illustrate the powerfulness of our approach by investigating the links between RNA-binding ability and other protein features, such as structural disorder and aggregation, in S. cerevisiae, C. elegans, M. musculus and H. sapiens. Our results are in striking agreement with available experimental evidence and unravel features that are key to understand the mechanisms regulating cellular homeostasis.
Conclusions: In an intuitive way, multiCM and cleverGO provide accurate classifications of physico-chemical features and annotations of biological processes, molecular functions and cellular components, which is extremely useful for the discovery and characterization of new trends in protein datasets. The multiCM and cleverGO can be freely accessed on the Web at http://www.tartaglialab.com/cs_multi/submission and http://www.tartaglialab.com/GO_analyser/universal . Each of the pages contains links to the corresponding documentation and tutorial.
Figures

References
-
- Klus P, Bolognesi B, Agostini F, Marchese D, Zanzoni A, Tartaglia GG. The cleverSuite Approach for Protein Characterization: Predictions of Structural Properties, Solubility, Chaperone Requirements and RNA-Binding Abilities. Bioinformatics. 2014;30(11):1601–8. doi: 10.1093/bioinformatics/btu074. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

