Maintenance of phenotypic diversity within a set of virulence encoding genes of the malaria parasite Plasmodium falciparum
- PMID: 26674193
- PMCID: PMC4707858
- DOI: 10.1098/rsif.2015.0848
Maintenance of phenotypic diversity within a set of virulence encoding genes of the malaria parasite Plasmodium falciparum
Abstract
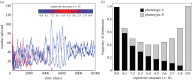
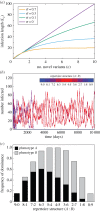
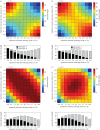
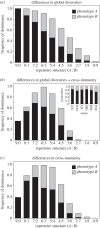
Infection by the human malaria parasite Plasmodium falciparum results in a broad spectrum of clinical outcomes, ranging from severe and potentially life-threatening malaria to asymptomatic carriage. In a process of naturally acquired immunity, individuals living in malaria-endemic regions build up a level of clinical protection, which attenuates infection severity in an exposure-dependent manner. Underlying this shift in the immunoepidemiology as well as the observed range in malaria pathogenesis is the var multigene family and the phenotypic diversity embedded within. The var gene-encoded surface proteins Plasmodium falciparum erythrocyte membrane protein 1 mediate variant-specific binding of infected red blood cells to a diverse set of host receptors that has been linked to specific disease manifestations, including cerebral and pregnancy-associated malaria. Here, we show that cross-reactive immune responses, which minimize the within-host benefit of each additionally expressed gene during infection, can cause selection for maximum phenotypic diversity at the genome level. We further show that differential functional constraints on protein diversification stably maintain uneven ratios between phenotypic groups, in line with empirical observation. Our results thus suggest that the maintenance of phenotypic diversity within P. falciparum is driven by an evolutionary trade-off that optimizes between within-host parasite fitness and between-host selection pressure.
Keywords: Plasmodium falciparum; evolutionary trade-off; mathematical modelling; phenotypic diversity; var genes.
© 2015 The Authors.
Figures
References
-
- Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82, 89–100. (10.1016/0092-8674(95)90055-1) - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
