H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes
- PMID: 26675384
- PMCID: PMC4698194
- DOI: 10.1016/j.tig.2015.11.001
H3K9me3-Dependent Heterochromatin: Barrier to Cell Fate Changes
Abstract
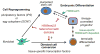
Establishing and maintaining cell identity depends on the proper regulation of gene expression, as specified by transcription factors and reinforced by epigenetic mechanisms. Among the epigenetic mechanisms, heterochromatin formation is crucial for the preservation of genome stability and the cell type-specific silencing of genes. The heterochromatin-associated histone mark H3K9me3, although traditionally associated with the noncoding portions of the genome, has emerged as a key player in repressing lineage-inappropriate genes and shielding them from activation by transcription factors. Here we describe the role of H3K9me3 heterochromatin in impeding the reprogramming of cell identity and the mechanisms by which H3K9me3 is reorganized during development and cell fate determination.
Keywords: H3K9me3; cell identity; development; heterochromatin; reprogramming.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

References
-
- Fisher AG, Merkenschlager M. Gene silencing, cell fate and nuclear organisation. Curr Opin Genet Dev. 2002;12:193–197. - PubMed
-
- Hemberger M, et al. Epigenetic dynamics of stem cells and cell lineage commitment: digging Waddington’s canal. Nat Rev Mol Cell Biol. 2009;10:526–537. - PubMed
-
- Heitz E. Das Heterochromatin der Moose. Bornträger; 1928.
-
- Wallrath LL, Elgin SC. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 1995;9:1263–1277. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

