miR-34a Silences c-SRC to Attenuate Tumor Growth in Triple-Negative Breast Cancer
- PMID: 26676753
- PMCID: PMC4755913
- DOI: 10.1158/0008-5472.CAN-15-2321
miR-34a Silences c-SRC to Attenuate Tumor Growth in Triple-Negative Breast Cancer
Abstract
Triple-negative breast cancer (TNBC) is an aggressive subtype with no clinically proven biologically targeted treatment options. The molecular heterogeneity of TNBC and lack of high frequency driver mutations other than TP53 have hindered the development of new and effective therapies that significantly improve patient outcomes. miRNAs, global regulators of survival and proliferation pathways important in tumor development and maintenance, are becoming promising therapeutic agents. We performed miRNA-profiling studies in different TNBC subtypes to identify miRNAs that significantly contribute to disease progression. We found that miR-34a was lost in TNBC, specifically within mesenchymal and mesenchymal stem cell-like subtypes, whereas expression of miR-34a targets was significantly enriched. Furthermore, restoration of miR-34a in cell lines representing these subtypes inhibited proliferation and invasion, activated senescence, and promoted sensitivity to dasatinib by targeting the proto-oncogene c-SRC. Notably, SRC depletion in TNBC cell lines phenocopied the effects of miR-34a reintroduction, whereas SRC overexpression rescued the antitumorigenic properties mediated by miR-34a. miR-34a levels also increased when cells were treated with c-SRC inhibitors, suggesting a negative feedback exists between miR-34a and c-SRC. Moreover, miR-34a administration significantly delayed tumor growth of subcutaneously and orthotopically implanted tumors in nude mice, and was accompanied by c-SRC downregulation. Finally, we found that miR-34a and SRC levels were inversely correlated in human tumor specimens. Together, our results demonstrate that miR-34a exerts potent antitumorigenic effects in vitro and in vivo and suggests that miR-34a replacement therapy, which is currently being tested in human clinical trials, represents a promising therapeutic strategy for TNBC.
©2015 American Association for Cancer Research.
Conflict of interest statement
Figures

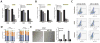
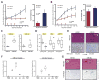


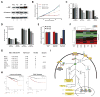
References
-
- Liedtke C, Gonzalez-Angulo A, Pusztai L. PPO Updat Princ Pract Oncol. 2. Vol. 24. New York: Lippincott Williams & Wilkins; 2010. Definition of triple-negative breast cancer and relationship to basal-like molecular subtype; pp. 1–6.
-
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–34. - PubMed
-
- Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol. 2006;24:5652–7. - PubMed
-
- Metzger-Filho O, Sun Z, Viale G, Price KN, Crivellari D, Snyder RD, et al. Patterns of Recurrence and Outcome According to Breast Cancer Subtypes in Lymph Node-Negative Disease: Results From International Breast Cancer Study Group Trials VIII and IX. J Clin Oncol. 2013 JCO.2012.46.1574–. - PMC - PubMed
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

