Human MicroRNA miR-532-5p Exhibits Antiviral Activity against West Nile Virus via Suppression of Host Genes SESTD1 and TAB3 Required for Virus Replication
- PMID: 26676784
- PMCID: PMC4810706
- DOI: 10.1128/JVI.02608-15
Human MicroRNA miR-532-5p Exhibits Antiviral Activity against West Nile Virus via Suppression of Host Genes SESTD1 and TAB3 Required for Virus Replication
Abstract
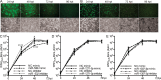
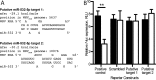
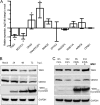
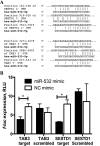
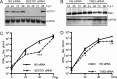
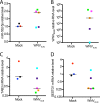
West Nile virus (WNV) is a mosquito-transmitted flavivirus that naturally circulates between mosquitos and birds but can also infect humans, causing severe neurological disease. The early host response to WNV infection in vertebrates primarily relies on the type I interferon pathway; however, recent studies suggest that microRNAs (miRNAs) may also play a notable role. In this study, we assessed the role of host miRNAs in response to WNV infection in human cells. We employed small RNA sequencing (RNA-seq) analysis to determine changes in the expression of host miRNAs in HEK293 cells infected with an Australian strain of WNV, Kunjin (WNVKUN), and identified a number of host miRNAs differentially expressed in response to infection. Three of these miRNAs were confirmed to be significantly upregulated in infected cells by quantitative reverse transcription (qRT)-PCR and Northern blot analyses, and one of them, miR-532-5p, exhibited a significant antiviral effect against WNVKUN infection. We have demonstrated that miR-532-5p targets and downregulates expression of the host genes SESTD1 and TAB3 in human cells. Small interfering RNA (siRNA) depletion studies showed that both SESTD1 and TAB3 were required for efficient WNVKUN replication. We also demonstrated upregulation of mir-532-5p expression and a corresponding decrease in the expression of its targets, SESTD1 and TAB3, in the brains of WNVKUN -infected mice. Our results show that upregulation of miR-532-5p and subsequent suppression of the SESTD1 and TAB3 genes represent a host antiviral response aimed at limiting WNVKUN infection and highlight the important role of miRNAs in controlling RNA virus infections in mammalian hosts.
Importance: West Nile virus (WNV) is a significant viral pathogen that poses a considerable threat to human health across the globe. There is no specific treatment or licensed vaccine available for WNV, and deeper insight into how the virus interacts with the host is required to facilitate their development. In this study, we addressed the role of host microRNAs (miRNAs) in antiviral response to WNV in human cells. We identified miR-532-5p as a novel antiviral miRNA and showed that it is upregulated in response to WNV infection and suppresses the expression of the host genes TAB3 and SESTD1 required for WNV replication. Our results show that upregulation of miR-532-5p and subsequent suppression of the SESTD1 and TAB3 genes represent an antiviral response aimed at limiting WNV infection and highlight the important role of miRNAs in controlling virus infections in mammalian hosts.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Hall RA, Broom AK, Smith DW, Mackenzie JS. 2002. The ecology and epidemiology of Kunjin virus. Curr Top Microbiol Immunol 267:253–269. - PubMed
-
- Frost MJ, Zhang J, Edmonds JH, Prow NA, Gu X, Davis R, Hornitzky C, Arzey KE, Finlaison D, Hick P, Read A, Hobson-Peters J, May FJ, Doggett SL, Haniotis J, Russell RC, Hall RA, Khromykh AA, Kirkland PD. 2012. Characterization of virulent West Nile virus Kunjin strain, Australia, 2011. Emerg Infect Dis 18:792–800. doi: 10.3201/eid1805.111720. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

