Nanobodies as Probes for Protein Dynamics in Vitro and in Cells
- PMID: 26677230
- PMCID: PMC4759159
- DOI: 10.1074/jbc.R115.679811
Nanobodies as Probes for Protein Dynamics in Vitro and in Cells
Abstract
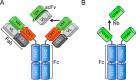
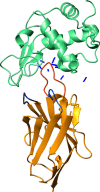
Nanobodies are the recombinant antigen-recognizing domains of the minimalistic heavy chain-only antibodies produced by camels and llamas. Nanobodies can be easily generated, effectively optimized, and variously derivatized with standard molecular biology protocols. These properties have triggered the recent explosion in the nanobody use in basic and clinical research. This review focuses on the emerging use of nanobodies for understanding and monitoring protein dynamics on the scales ranging from isolated protein domains to live cells, from nanoseconds to hours. The small size and high solubility make nanobodies uniquely suited for studying protein dynamics by NMR. The ability to produce conformation-sensitive nanobodies in cells enables studies that link structural dynamics of a target protein to its cellular behavior. The link between in vitro and in-cell dynamics, afforded by nanobodies, brings the analysis of such important events as receptor signaling, membrane protein trafficking, and protein interactions to the next level of resolution.
Keywords: X-ray crystallography; nanobody; nuclear magnetic resonance (NMR); protein domain; protein dynamic; protein dynamics; protein engineering; protein structure; single-domain antibody (sdAb, nanobody).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E. B., Bendahman N., and Hamers R. (1993) Naturally occurring antibodies devoid of light chains. Nature 363, 446–448 - PubMed
-
- Broisat A., Hernot S., Toczek J., De Vos J., Riou L. M., Martin S., Ahmadi M., Thielens N., Wernery U., Caveliers V., Muyldermans S., Lahoutte T., Fagret D., Ghezzi C., and Devoogdt N. (2012) Nanobodies targeting mouse/human VCAM1 for the nuclear imaging of atherosclerotic lesions. Circ. Res. 110, 927–937 - PMC - PubMed
-
- Caussinus E., Kanca O., and Affolter M. (2012) Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol. 19, 117–121 - PubMed
-
- Rothbauer U., Zolghadr K., Tillib S., Nowak D., Schermelleh L., Gahl A., Backmann N., Conrath K., Muyldermans S., Cardoso M. C., and Leonhardt H. (2006) Targeting and tracing antigens in live cells with fluorescent nanobodies. Nat. Methods 3, 887–889 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

