Performance of Copan WASP for Routine Urine Microbiology
- PMID: 26677255
- PMCID: PMC4767997
- DOI: 10.1128/JCM.02577-15
Performance of Copan WASP for Routine Urine Microbiology
Abstract
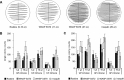
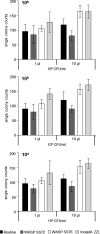
This study compared a manual workup of urine clinical samples with fully automated WASPLab processing. As a first step, two different inocula (1 and 10 μl) and different streaking patterns were compared using WASP and InoqulA BT instrumentation. Significantly more single colonies were produced with the10-μl inoculum than with the 1-μl inoculum, and automated streaking yielded significantly more single colonies than manual streaking on whole plates (P < 0.001). In a second step, 379 clinical urine samples were evaluated using WASP and the manual workup. Average numbers of detected morphologies, recovered species, and CFUs per milliliter of all 379 urine samples showed excellent agreement between WASPLab and the manual workup. The percentage of urine samples clinically categorized as positive or negative did not differ between the automated and manual workflow, but within the positive samples, automated processing by WASPLab resulted in the detection of more potential pathogens. In summary, the present study demonstrates that (i) the streaking pattern, i.e., primarily the number of zigzags/length of streaking lines, is critical for optimizing the number of single colonies yielded from primary cultures of urine samples; (ii) automated streaking by the WASP instrument is superior to manual streaking regarding the number of single colonies yielded (for 32.2% of the samples); and (iii) automated streaking leads to higher numbers of detected morphologies (for 47.5% of the samples), species (for 17.4% of the samples), and pathogens (for 3.4% of the samples). The results of this study point to an improved quality of microbiological analyses and laboratory reports when using automated sample processing by WASP and WASPLab.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

