Using activation status of signaling pathways as mechanism-based biomarkers to predict drug sensitivity
- PMID: 26678097
- PMCID: PMC4683444
- DOI: 10.1038/srep18494
Using activation status of signaling pathways as mechanism-based biomarkers to predict drug sensitivity
Abstract
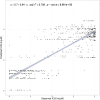
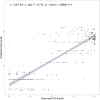
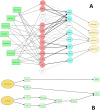
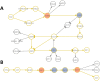
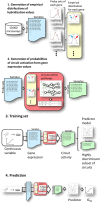
Many complex traits, as drug response, are associated with changes in biological pathways rather than being caused by single gene alterations. Here, a predictive framework is presented in which gene expression data are recoded into activity statuses of signal transduction circuits (sub-pathways within signaling pathways that connect receptor proteins to final effector proteins that trigger cell actions). Such activity values are used as features by a prediction algorithm which can efficiently predict a continuous variable such as the IC50 value. The main advantage of this prediction method is that the features selected by the predictor, the signaling circuits, are themselves rich-informative, mechanism-based biomarkers which provide insight into or drug molecular mechanisms of action (MoA).
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

