Liver transcriptomic networks reveal main biological processes associated with feed efficiency in beef cattle
- PMID: 26678995
- PMCID: PMC4683712
- DOI: 10.1186/s12864-015-2292-8
Liver transcriptomic networks reveal main biological processes associated with feed efficiency in beef cattle
Erratum in
-
Erratum to: Liver transcriptomic networks reveal main biological processes associated with feed efficiency in beef cattle.BMC Genomics. 2016 Apr 28;17:311. doi: 10.1186/s12864-016-2649-7. BMC Genomics. 2016. PMID: 27126189 Free PMC article. No abstract available.
-
Erratum to: 'Reference-free inference of tumor phylogenies from single-cell sequencing data'.BMC Genomics. 2016 May 10;17(1):348. doi: 10.1186/s12864-016-2609-2. BMC Genomics. 2016. PMID: 27164840 Free PMC article. No abstract available.
Abstract
Background: The selection of beef cattle for feed efficiency (FE) traits is very important not only for productive and economic efficiency but also for reduced environmental impact of livestock. Considering that FE is multifactorial and expensive to measure, the aim of this study was to identify biological functions and regulatory genes associated with this phenotype.
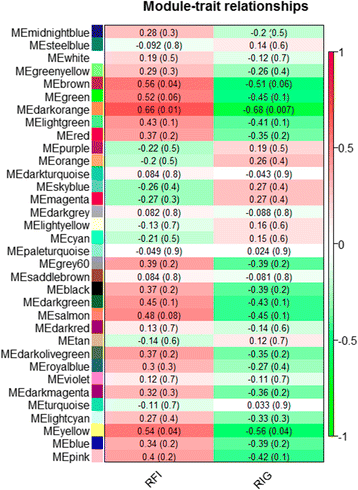
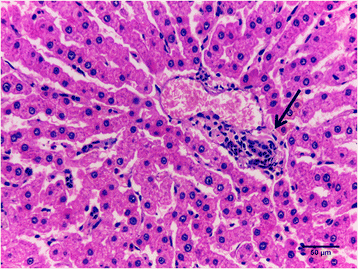
Results: Eight genes were differentially expressed between high and low feed efficient animals (HFE and LFE, respectively). Co-expression analyses identified 34 gene modules of which 4 were strongly associated with FE traits. They were mainly enriched for inflammatory response or inflammation-related terms. We also identified 463 differentially co-expressed genes which were functionally enriched for immune response and lipid metabolism. A total of 8 key regulators of gene expression profiles affecting FE were found. The LFE animals had higher feed intake and increased subcutaneous and visceral fat deposition. In addition, LFE animals showed higher levels of serum cholesterol and liver injury biomarker GGT. Histopathology of the liver showed higher percentage of periportal inflammation with mononuclear infiltrate.
Conclusion: Liver transcriptomic network analysis coupled with other results demonstrated that LFE animals present altered lipid metabolism and increased hepatic periportal lesions associated with an inflammatory response composed mainly by mononuclear cells. We are now focusing to identify the causes of increased liver lesions in LFE animals.
Figures
References
-
- United Nations, Department of Economic and Social Affairs P division. World Population Prospects: The 2010 Revision, Highlights and Advance Tables. New York; 2011. http://www.un.org/en/development/desa/population/publications/pdf/trends....
-
- Arthur PF, Archer JA, Herd RM. Feed intake and efficiency in beef cattle: overview of recent Australian research and challenges for the future. Aust J Exp Agric. 2004;44:361. doi: 10.1071/EA02162. - DOI
-
- Gerber PJ, Steinfeld H, Henderson B, Mottet A, Opio C, Dijkman J, et al. Tackling Climate Change Through Livestock - A Global Assessment of Emissions and Mitigation Opportunities. 2013.
-
- Nkrumah JD, Okine EK, Mathison GW, Schmid K, Li C, Basarab JA, et al. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J Anim Sci. 2006;84:145–53. - PubMed
-
- Koch RM, Swiger LA, Chambers D, Gregory KE. Efficiency of Feed Use in Beef Cattle. J Anim Sci. 1963;22:486–494.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous