Three easy pieces
- PMID: 26679422
- PMCID: PMC4799745
- DOI: 10.1016/j.bbagen.2015.12.003
Three easy pieces
Abstract
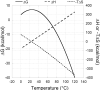
Background: Differential scanning calorimetry is a powerful method that provides a complete thermodynamic characterization of the stability of a protein as a function of temperature. There are, however, circumstances that preclude a complete analysis of DSC data. The most common ones are irreversible denaturation transitions or transitions that take place at temperatures that are beyond the temperature limit of the instrument. Even for a protein that undergoes reversible thermal denaturation, the extrapolation of the thermodynamic data to lower temperatures, usually 25°C, may become unreliable due to difficulties in the determination of ΔCp.
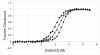
Methods: The combination of differential scanning calorimetry and isothermal chemical denaturation allows reliable thermodynamic analysis of protein stability under less than ideal conditions.
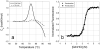
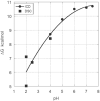
Results and conclusions: This paper demonstrates how DSC can be used in combination with chemical denaturation to address three different scenarios: 1) estimation of an accurate ΔCp value for a reversible denaturation using as a test system the envelope HIV-1 glycoprotein gp120; 2) determination of the Gibbs energy of stability in the region in which thermal denaturation is irreversible using HEW lysozyme at different pH values; and, 3) determination of Gibbs energy of stability for a thermostable protein, thermolysin.
Keywords: Chemical denaturation; DSC; HEW lysozyme; HIV-1 gp120; ICD; Thermolysin.
Copyright © 2015 Elsevier B.V. All rights reserved.
Figures





References
-
- Brandts JF, Oliveira RJ, Westort C. Thermodynamics of protein denaturation. Effect of pressure on the denaturation of ribonuclease A. Biochemistry. 1970;9:1038–1047. - PubMed
-
- Privalov G, Kavina V, Freire E, Privalov PL. Precise scanning calorimeter for studying thermal properties of biological macromolecules in dilute solution. Anal Biochem. 1995;232:79–85. - PubMed
-
- Privalov PL, Khechinashvili NN. A thermodynamic approach to the problem of stabilization of globular protein structure: a calorimetric study. J Mol Biol. 1974;86:665–684. - PubMed
-
- Freire E, Biltonen RL. Statistical mechanical deconvolution of thermal transitions in macromolecules. I. Theory and application to homogeneous systems. Biopolymers. 1978;17:463–479.
-
- Privalov PL, Khechinashvili NN, Atanasov BP. Thermodynamic analysis of thermal transitions in globular proteins. I. Calorimetric study of chymotrypsinogen, ribonuclease and myoglobin. Biopolymers. 1971;10:1865–1890. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

