Structural Correlation of the Neck Coil with the Coiled-coil (CC1)-Forkhead-associated (FHA) Tandem for Active Kinesin-3 KIF13A
- PMID: 26680000
- PMCID: PMC4751397
- DOI: 10.1074/jbc.M115.689091
Structural Correlation of the Neck Coil with the Coiled-coil (CC1)-Forkhead-associated (FHA) Tandem for Active Kinesin-3 KIF13A
Abstract
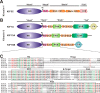
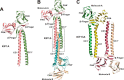
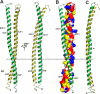
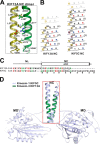
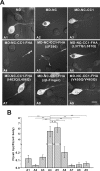
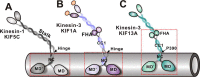
Processive kinesin motors often contain a coiled-coil neck that controls the directionality and processivity. However, the neck coil (NC) of kinesin-3 is too short to form a stable coiled-coil dimer. Here, we found that the coiled-coil (CC1)-forkhead-associated (FHA) tandem (that is connected to NC by Pro-390) of kinesin-3 KIF13A assembles as an extended dimer. With the removal of Pro-390, the NC-CC1 tandem of KIF13A unexpectedly forms a continuous coiled-coil dimer that can be well aligned into the CC1-FHA dimer. The reverse introduction of Pro-390 breaks the NC-CC1 coiled-coil dimer but provides the intrinsic flexibility to couple NC with the CC1-FHA tandem. Mutations of either NC, CC1, or the FHA domain all significantly impaired the motor activity. Thus, the three elements within the NC-CC1-FHA tandem of KIF13A are structurally interrelated to form a stable dimer for activating the motor. This work also provides the first direct structural evidence to support the formation of a coiled-coil neck by the short characteristic neck domain of kinesin-3.
Keywords: KIF13A; intracellular trafficking; kinesin; molecular motor; neck coil; structural biology; x-ray crystallography.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Vale R. D. (2003) The molecular motor toolbox for intracellular transport. Cell 112, 467–480 - PubMed
-
- Soldati T., and Schliwa M. (2006) Powering membrane traffic in endocytosis and recycling. Nat. Rev. Mol. Cell Biol. 7, 897–908 - PubMed
-
- Hirokawa N., Niwa S., and Tanaka Y. (2010) Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron 68, 610–638 - PubMed
-
- Hirokawa N., Noda Y., Tanaka Y., and Niwa S. (2009) Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 10, 682–696 - PubMed
-
- Verhey K. J., Kaul N., and Soppina V. (2011) Kinesin assembly and movement in cells. Annu. Rev. Biophys. 40, 267–288 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

