Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing
- PMID: 26681312
- PMCID: PMC4985612
- DOI: 10.1038/gim.2015.166
Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing
Erratum in
-
CORRIGENDUM: Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing.Genet Med. 2016 May;18(5):531-2. doi: 10.1038/gim.2016.21. Genet Med. 2016. PMID: 27126497 Free PMC article. No abstract available.
Abstract
Purpose: Germ-line testing for panels of cancer genes using next-generation sequencing is becoming more common in clinical care. We report our experience as a clinical laboratory testing both well-established, high-risk cancer genes (e.g., BRCA1/2, MLH1, MSH2) as well as more recently identified cancer genes (e.g., PALB2, BRIP1), many of which have increased but less well-defined penetrance.
Methods: Clinical genetic testing was performed on over 10,000 consecutive cases referred for evaluation of germ-line cancer genes, and results were analyzed for frequency of pathogenic or likely pathogenic variants, and were stratified by testing panel, gene, and clinical history.
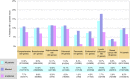
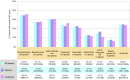
Results: Overall, a molecular diagnosis was made in 9.0% of patients tested, with the highest yield in the Lynch syndrome/colorectal cancer panel. In patients with breast, ovarian, or colon/stomach cancer, positive yields were 9.7, 13.4, and 14.8%, respectively. Approximately half of the pathogenic variants identified in patients with breast or ovarian cancer were in genes other than BRCA1/2.
Conclusion: The high frequency of positive results in a wide range of cancer genes, including those of high penetrance and with clinical care guidelines, underscores both the genetic heterogeneity of hereditary cancer and the usefulness of multigene panels over genetic tests of one or two genes.Genet Med 18 8, 823-832.
Figures
References
-
- Tung N, Battelli C, Allen B, et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer 2015;121:25–33. - PubMed
-
- Richards CS, Bale S, Bellissimo DB, et al.; Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med 2008;10:294–300. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

