Molecular control of activation and priming in macrophages
- PMID: 26681459
- PMCID: PMC4795476
- DOI: 10.1038/ni.3306
Molecular control of activation and priming in macrophages
Abstract
In tissues, macrophages are exposed to metabolic, homeostatic and immunoregulatory signals of local or systemic origin that influence their basal functions and responses to danger signals. Signal-transduction pathways regulated by extracellular signals are coupled to distinct sets of broadly expressed stimulus-regulated transcription factors whose ability to elicit gene-expression changes is influenced by the accessibility of their binding sites in the macrophage genome. In turn, accessibility of macrophage-specific transcriptional regulatory elements (enhancers and promoters) is specified by transcription factors that determine the macrophage lineage or impose their tissue-specific properties. Here we review recent findings that advance the understanding of mechanisms underlying priming and signal-dependent activation of macrophages and discuss the effect of genetic variation on these processes.
Figures


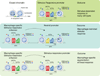

References
-
- Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nature reviews. Immunology. 2011;11:750–761. - PubMed
-
- Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harbor symposia on quantitative biology. 1989;54(Pt 1):1–13. - PubMed
-
- Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annual review of immunology. 2002;20:197–216. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

