Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons
- PMID: 26681692
- PMCID: PMC4737166
- DOI: 10.1093/nar/gkv1470
Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons
Abstract
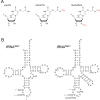
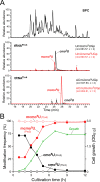
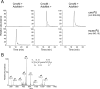
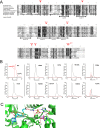
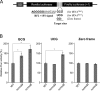
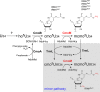
Post-transcriptional modifications at the anticodon first (wobble) position of tRNA play critical roles in precise decoding of genetic codes. 5-carboxymethoxyuridine (cmo(5)U) and its methyl ester derivative 5-methoxycarbonylmethoxyuridine (mcmo(5)U) are modified nucleosides found at the anticodon wobble position in several tRNAs from Gram-negative bacteria. cmo(5)U and mcmo(5)U facilitate non-Watson-Crick base pairing with guanosine and pyrimidines at the third positions of codons, thereby expanding decoding capabilities. By mass spectrometric analyses of individual tRNAs and a shotgun approach of total RNA from Escherichia coli, we identified mcmo(5)U as a major modification in tRNA(Ala1), tRNA(Ser1), tRNA(Pro3) and tRNA(Thr4); by contrast, cmo(5)U was present primarily in tRNA(Leu3) and tRNA(Val1). In addition, we discovered 5-methoxycarbonylmethoxy-2'-O-methyluridine (mcmo(5)Um) as a novel but minor modification in tRNA(Ser1). Terminal methylation frequency of mcmo(5)U in tRNA(Pro3) was low (≈30%) in the early log phase of cell growth, gradually increased as growth proceeded and reached nearly 100% in late log and stationary phases. We identified CmoM (previously known as SmtA), an AdoMet-dependent methyltransferase that methylates cmo(5)U to form mcmo(5)U. A luciferase reporter assay based on a +1 frameshift construct revealed that terminal methylation of mcmo(5)U contributes to the decoding ability of tRNA(Ala1).
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Yokoyama S., Nishimura S. In: tRNA: Structure, Biosynthesis, and Function. Soll DRUL, editor. Washington, D.C.: American Society for Microbiology; 1995. pp. 207–224.
-
- Bjork G. In: tRNA: Structure, Biosynthesis, and Function. Soll DRUL, editor. Washington, D.C.: American Society for Microbiology; 1995. pp. 165–205.
-
- Suzuki T. In: Fine-Tuning of RNA Functions by Modification and Editing. Grosjean H, editor. Vol. 12. Springer-Verlag Berlin and Heidelberg: GmbH & Co. KG; 2005. pp. 23–69.
-
- Crick F.H. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 1966;19:548–555. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

