Targeted next-generation sequencing panel (GlioSeq) provides comprehensive genetic profiling of central nervous system tumors
- PMID: 26681766
- PMCID: PMC4767245
- DOI: 10.1093/neuonc/nov289
Targeted next-generation sequencing panel (GlioSeq) provides comprehensive genetic profiling of central nervous system tumors
Abstract
Background: Identification of genetic changes in CNS tumors is important for the appropriate clinical management of patients. Our objective was to develop a next-generation sequencing (NGS) assay for simultaneously detecting the various types of genetic alterations characteristic for adult and pediatric CNS tumors that can be applied to small brain biopsies.
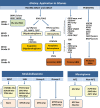
Methods: We report an amplification-based targeted NGS assay (GlioSeq) that analyzes 30 genes for single nucleotide variants (SNVs) and indels, 24 genes for copy number variations (CNVs), and 14 types of structural alterations in BRAF, EGFR, and FGFR3 genes in a single workflow. GlioSeq performance was evaluated in 54 adult and pediatric CNS tumors, and the results were compared with fluorescence in-situ hybridization, Sanger sequencing, and reverse transcription PCR.
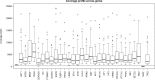
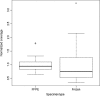
Results: GlioSeq correctly identified 71/71 (100%) genetic alterations known to be present by conventional techniques, including 56 SNVs/indels, 9 CNVs, 3 EGFRvIII, and 3 KIAA1549-BRAF fusions. Only 20 ng of DNA and 10 ng of RNA were required for successful sequencing of 100% frozen and 96% formalin-fixed, paraffin-embedded tissue specimens. The assay sensitivity was 3%-5% of mutant alleles for SNVs and 1%-5% for gene fusions. The most commonly detected alterations were IDH1, TP53, TERT, ATRX. CDKN2A, and PTEN in high-grade gliomas, followed by BRAF fusions in low-grade gliomas and H3F3A mutations in pediatric gliomas.
Conclusions: GlioSeq NGS assay offers accurate and sensitive detection of a wide range of genetic alterations in a single workflow. It allows rapid and cost-effective profiling of brain tumor specimens and thus provides valuable information for patient management.
Keywords: CNS tumors; gene fusions; mutations; next generation sequencing; paraffin.
© The Author(s) 2015. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


Comment in
-
Next-generation sequencing of central nervous systems tumors: the future of personalized patient management.Neuro Oncol. 2016 Mar;18(3):308-10. doi: 10.1093/neuonc/nov329. Neuro Oncol. 2016. PMID: 26917589 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

