To Know How a Gene Works, We Need to Redefine It First but then, More Importantly, to Let the Cell Itself Decide How to Transcribe and Process Its RNAs
- PMID: 26681921
- PMCID: PMC4671999
- DOI: 10.7150/ijbs.13436
To Know How a Gene Works, We Need to Redefine It First but then, More Importantly, to Let the Cell Itself Decide How to Transcribe and Process Its RNAs
Abstract
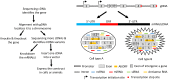
Recent genomic and ribonomic research reveals that our genome produces a stupendous amount of non-coding RNAs (ncRNAs), including antisense RNAs, and that many genes contain other gene(s) in their introns. Since ncRNAs either regulate the transcription, translation or stability of mRNAs or directly exert cellular functions, they should be regarded as the fourth category of RNAs, after ribosomal, messenger and transfer RNAs. These and other research advances challenge the current concept of gene and raise a question as to how we should redefine gene. We can either consider each tiny part of the classically-defined gene, such as each mRNA variant, as a "gene", or, alternatively and oppositely, regard a whole genomic locus as a "gene" that may contain intron-embedded genes and produce different types of RNAs and proteins. Each of the two ways to redefine gene not only has its strengths and weaknesses but also has its particular concern on the methodology for the determination of the gene's function: Ectopic expression of complementary DNA (cDNA) in cells has in the past decades provided us with great deal of detail about the functions of individual mRNA variants, and will make the data less conflicting with each other if just a small part of a classically-defined gene is considered as a "gene". On the other hand, genomic DNA (gDNA) will better help us in understanding the collective function of a genomic locus. In our opinion, we need to be more cautious in the use of cDNA and in the explanation of data resulting from cDNA, and, instead, should make delivery of gDNA into cells routine in determination of genes' functions, although this demands some technology renovation.
Keywords: Complementary DNA; Gene definition; Gene function; Genomic locus; non-coding RNA.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



Similar articles
-
[Analysis, identification and correction of some errors of model refseqs appeared in NCBI Human Gene Database by in silico cloning and experimental verification of novel human genes].Yi Chuan Xue Bao. 2004 May;31(5):431-43. Yi Chuan Xue Bao. 2004. PMID: 15478601 Chinese.
-
Noncoding RNA transcripts.J Appl Genet. 2003;44(1):1-19. J Appl Genet. 2003. PMID: 12590177 Review.
-
Life without RNAi: noncoding RNAs and their functions in Saccharomyces cerevisiae.Biochem Cell Biol. 2009 Oct;87(5):767-79. doi: 10.1139/O09-043. Biochem Cell Biol. 2009. PMID: 19898526 Review.
-
[Use of the real-time RT-PCR method for investigation of small stable RNA expression level in human epidermoid carcinoma cells A431].Tsitologiia. 2003;45(4):392-402. Tsitologiia. 2003. PMID: 14520871 Russian.
-
Multiple RNAs from the mouse carboxypeptidase M locus: functional RNAs or transcription noise?BMC Mol Biol. 2009 Feb 8;10:7. doi: 10.1186/1471-2199-10-7. BMC Mol Biol. 2009. PMID: 19200403 Free PMC article.
Cited by
-
It Is Imperative to Establish a Pellucid Definition of Chimeric RNA and to Clear Up a Lot of Confusion in the Relevant Research.Int J Mol Sci. 2017 Mar 28;18(4):714. doi: 10.3390/ijms18040714. Int J Mol Sci. 2017. PMID: 28350330 Free PMC article. Review.
-
Mutation or not, what directly establishes a neoplastic state, namely cellular immortality and autonomy, still remains unknown and should be prioritized in our research.J Cancer. 2022 Jul 4;13(9):2810-2843. doi: 10.7150/jca.72628. eCollection 2022. J Cancer. 2022. PMID: 35912015 Free PMC article. Review.
-
ACTB and GAPDH appear at multiple SDS-PAGE positions, thus not suitable as reference genes for determining protein loading in techniques like Western blotting.Open Life Sci. 2021 Dec 13;16(1):1278-1292. doi: 10.1515/biol-2021-0130. eCollection 2021. Open Life Sci. 2021. PMID: 34966852 Free PMC article.
-
At elevated temperatures, heat shock protein genes show altered ratios of different RNAs and expression of new RNAs, including several novel HSPB1 mRNAs encoding HSP27 protein isoforms.Exp Ther Med. 2021 Aug;22(2):900. doi: 10.3892/etm.2021.10332. Epub 2021 Jun 24. Exp Ther Med. 2021. PMID: 34257713 Free PMC article.
-
The well-accepted notion that gene amplification contributes to increased expression still remains, after all these years, a reasonable but unproven assumption.J Carcinog. 2016 May 20;15:3. doi: 10.4103/1477-3163.182809. eCollection 2016. J Carcinog. 2016. PMID: 27298590 Free PMC article. Review.
References
-
- Zhang J, Lou X, Shen H, Zellmer L, Sun Y, Liu S. et al. Isoforms of wild type proteins often appear as low molecular weight bands on SDS-PAGE. Biotechnol J. 2014;9(8):1044–1054. - PubMed
-
- Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science. 2012;337(6099):1159. 1161-doi: 10.1126/science. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

