The Nucleotide-Binding Sites of SUR1: A Mechanistic Model
- PMID: 26682803
- PMCID: PMC4699857
- DOI: 10.1016/j.bpj.2015.10.026
The Nucleotide-Binding Sites of SUR1: A Mechanistic Model
Abstract
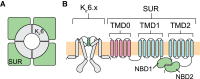
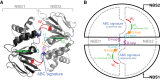
ATP-sensitive potassium (KATP) channels comprise four pore-forming Kir6.2 subunits and four modulatory sulfonylurea receptor (SUR) subunits. The latter belong to the ATP-binding cassette family of transporters. KATP channels are inhibited by ATP (or ADP) binding to Kir6.2 and activated by Mg-nucleotide interactions with SUR. This dual regulation enables the KATP channel to couple the metabolic state of a cell to its electrical excitability and is crucial for the KATP channel's role in regulating insulin secretion, cardiac and neuronal excitability, and vascular tone. Here, we review the regulation of the KATP channel by adenine nucleotides and present an equilibrium allosteric model for nucleotide activation and inhibition. The model can account for many experimental observations in the literature and provides testable predictions for future experiments.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Proks P., Ashcroft F.M. Modeling K(ATP) channel gating and its regulation. Prog. Biophys. Mol. Biol. 2009;99:7–19. - PubMed
-
- Nichols C.G., Shyng S.L., Bryan J. Adenosine diphosphate as an intracellular regulator of insulin secretion. Science. 1996;272:1785–1787. - PubMed
-
- Tucker S.J., Gribble F.M., Ashcroft F.M. Truncation of Kir6.2 produces ATP-sensitive K+ channels in the absence of the sulphonylurea receptor. Nature. 1997;387:179–183. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

