Contribution of the Salmonella enterica KdgR Regulon to Persistence of the Pathogen in Vegetable Soft Rots
- PMID: 26682862
- PMCID: PMC4751823
- DOI: 10.1128/AEM.03355-15
Contribution of the Salmonella enterica KdgR Regulon to Persistence of the Pathogen in Vegetable Soft Rots
Abstract
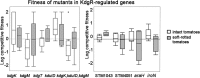
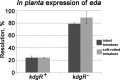
During their colonization of plants, human enteric pathogens, such as Salmonella enterica, are known to benefit from interactions with phytopathogens. At least in part, benefits derived by Salmonella from the association with a soft rot caused by Pectobacterium carotovorum were shown to be dependent on Salmonella KdgR, a regulator of genes involved in the uptake and utilization of carbon sources derived from the degradation of plant polymers. A Salmonella kdgR mutant was more fit in soft rots but not in the lesions caused by Xanthomonas spp. and Pseudomonas spp. Bioinformatic, phenotypic, and gene expression analyses demonstrated that the KdgR regulon included genes involved in uptake and metabolism of molecules resulting from pectin degradation as well as those central to the utilization of a number of other carbon sources. Mutant analyses indicated that the Entner-Doudoroff pathway, in part controlled by KdgR, was critical for the persistence within soft rots and likely was responsible for the kdgR phenotype.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

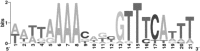



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

