Dysregulated miR-671-5p / CDR1-AS / CDR1 / VSNL1 axis is involved in glioblastoma multiforme
- PMID: 26683098
- PMCID: PMC4826240
- DOI: 10.18632/oncotarget.6621
Dysregulated miR-671-5p / CDR1-AS / CDR1 / VSNL1 axis is involved in glioblastoma multiforme
Abstract
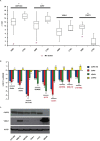
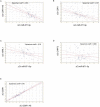
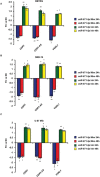
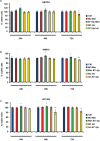
MiR-671-5p is encoded by a gene localized at 7q36.1, a region amplified in human glioblastoma multiforme (GBM), the most malignant brain cancer. To investigate whether expression of miR-671-5p were altered in GBM, we analyzed biopsies from a cohort of forty-five GBM patients and from five GBM cell lines. Our data show significant overexpression of miR-671-5p in both biopsies and cell lines. By exploiting specific miRNA mimics and inhibitors, we demonstrated that miR-671-5p overexpression significantly increases migration and to a less extent proliferation rates of GBM cells. Through a combined in silico and in vitro approach, we identified CDR1-AS, CDR1, VSNL1 as downstream miR-671-5p targets in GBM. Expression of these genes significantly decreased both in GBM biopsies and cell lines and negatively correlated with that of miR-671-5p. Based on our data, we propose that the axis miR-671-5p / CDR1-AS / CDR1 / VSNL1 is functionally altered in GBM cells and is involved in the modification of their biopathological profile.
Keywords: cell networks; circular RNAs (circRNAs); glioblastoma multiforme (GBM); microRNAs (miRNAs); non coding RNAs (ncRNAs).
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

