The Eukaryotic Mismatch Recognition Complexes Track with the Replisome during DNA Synthesis
- PMID: 26684201
- PMCID: PMC4684283
- DOI: 10.1371/journal.pgen.1005719
The Eukaryotic Mismatch Recognition Complexes Track with the Replisome during DNA Synthesis
Abstract
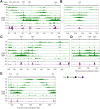
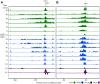
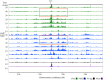
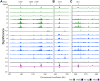
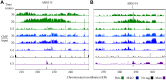
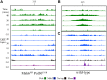
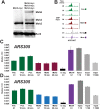
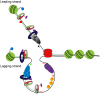
During replication, mismatch repair proteins recognize and repair mispaired bases that escape the proofreading activity of DNA polymerase. In this work, we tested the model that the eukaryotic mismatch recognition complex tracks with the advancing replisome. Using yeast, we examined the dynamics during replication of the leading strand polymerase Polε using Pol2 and the eukaryotic mismatch recognition complex using Msh2, the invariant protein involved in mismatch recognition. Specifically, we synchronized cells and processed samples using chromatin immunoprecipitation combined with custom DNA tiling arrays (ChIP-chip). The Polε signal was not detectable in G1, but was observed at active origins and replicating DNA throughout S-phase. The Polε signal provided the resolution to track origin firing timing and efficiencies as well as replisome progression rates. By detecting Polε and Msh2 dynamics within the same strain, we established that the mismatch recognition complex binds origins and spreads to adjacent regions with the replisome. In mismatch repair defective PCNA mutants, we observed that Msh2 binds to regions of replicating DNA, but the distribution and dynamics are altered, suggesting that PCNA is not the sole determinant for the mismatch recognition complex association with replicating regions, but may influence the dynamics of movement. Using biochemical and genomic methods, we provide evidence that both MutS complexes are in the vicinity of the replisome to efficiently repair the entire spectrum of mutations during replication. Our data supports the model that the proximity of MutSα/β to the replisome for the efficient repair of the newly synthesized strand before chromatin reassembles.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

