Molecular Characterization of Two Lysophospholipid:acyl-CoA Acyltransferases Belonging to the MBOAT Family in Nicotiana benthamiana
- PMID: 26684752
- PMCID: PMC4684200
- DOI: 10.1371/journal.pone.0144653
Molecular Characterization of Two Lysophospholipid:acyl-CoA Acyltransferases Belonging to the MBOAT Family in Nicotiana benthamiana
Abstract
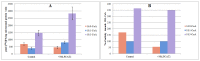
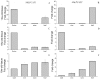
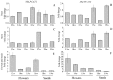
In the remodeling pathway for the synthesis of phosphatidylcholine (PC), acyl-CoA-dependent lysophosphatidylcholine (lysoPC) acyltransferase (LPCAT) catalyzes the reacylation of lysoPC. A number of genes encoding LPCATs have been cloned and characterized from several plants in recent years. Using Arabidopsis and other plant LPCAT sequences to screen the genome database of Nicotiana benthamiana, we identified two cDNAs encoding the putative tobacco LPCATs (NbLPCAT1 and NbLPCAT2). Both of them were predicted to encode a protein of 463 amino acids with high similarity to LPCATs from other plants. Protein sequence features such as the presence of at least eight putative transmembrane regions, four highly conserved signature motifs and several invariant residues indicate that NbLPCATs belong to the membrane bound O-acyltransferase family. Lysophospholipid acyltransferase activity of NbLPCATs was confirmed by testing lyso-platelet-activating factor (lysoPAF) sensitivity through heterologous expression of each full-length cDNA in a yeast mutant Y02431 (lca1△) disrupted in endogenous LPCAT enzyme activity. Analysis of fatty acid profiles of phospholipids from the NbLPCAT-expressing yeast mutant Y02431 cultures supplemented with polyunsaturated fatty acids suggested more incorporation of linoleic acid (18:2n6, LA) and α-linolenic acid (18:3n3, ALA) into PC compared to yeast mutant harbouring empty vector. In vitro enzymatic assay demonstrated that NbLPCAT1had high lysoPC acyltransferase activity with a clear preference for α-linolenoyl-CoA (18:3), while NbLPCAT2 showed a high lysophosphatidic acid (lysoPA) acyltransferase activity towards α-linolenoyl-CoA and a weak lysoPC acyltransferase activity. Tissue-specific expression analysis showed a ubiquitous expression of NbLPCAT1 and NbLPCAT2 in roots, stems, leaves, flowers and seeds, and a strong expression in developing flowers. This is the first report on the cloning and characterization of lysophospholipid acyltransferases from N. benthamiana.
Conflict of interest statement
Figures








References
-
- Shimizu T, Ohto T, Kita Y. Cytosolic phospholipase A2: Biochemical properties and physiological roles. IUBMB Life 2006; 58:328–333. - PubMed
-
- Kennedy EP, Weiss SB. The function of cytidine coenzymes in the biosynthesis of phospholipids. J Biol Chem. 1956; 222: 193–214. - PubMed
-
- Lands WE. Metabolism of glycerolipids. 2. The enzymatic acylation of lysolecithin. J Biol Chem. 1960; 235: 2233–2237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

