Deformable MR Prostate Segmentation via Deep Feature Learning and Sparse Patch Matching
- PMID: 26685226
- PMCID: PMC5002995
- DOI: 10.1109/TMI.2015.2508280
Deformable MR Prostate Segmentation via Deep Feature Learning and Sparse Patch Matching
Abstract
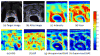
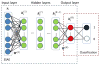
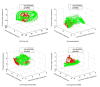
Automatic and reliable segmentation of the prostate is an important but difficult task for various clinical applications such as prostate cancer radiotherapy. The main challenges for accurate MR prostate localization lie in two aspects: (1) inhomogeneous and inconsistent appearance around prostate boundary, and (2) the large shape variation across different patients. To tackle these two problems, we propose a new deformable MR prostate segmentation method by unifying deep feature learning with the sparse patch matching. First, instead of directly using handcrafted features, we propose to learn the latent feature representation from prostate MR images by the stacked sparse auto-encoder (SSAE). Since the deep learning algorithm learns the feature hierarchy from the data, the learned features are often more concise and effective than the handcrafted features in describing the underlying data. To improve the discriminability of learned features, we further refine the feature representation in a supervised fashion. Second, based on the learned features, a sparse patch matching method is proposed to infer a prostate likelihood map by transferring the prostate labels from multiple atlases to the new prostate MR image. Finally, a deformable segmentation is used to integrate a sparse shape model with the prostate likelihood map for achieving the final segmentation. The proposed method has been extensively evaluated on the dataset that contains 66 T2-wighted prostate MR images. Experimental results show that the deep-learned features are more effective than the handcrafted features in guiding MR prostate segmentation. Moreover, our method shows superior performance than other state-of-the-art segmentation methods.
Figures













References
-
- Prostate Cancer. Available: http://www.cancer.org/acs/groups/cid/documents/webcontent/003134-pdf.pdf.
-
- Pondman KM, Fütterer JJ, ten Haken B, Schultze Kool LJ, Witjes JA, Hambrock T, et al. MR-guided biopsy of the prostate: an overview of techniques and a systematic review. European Urology. 2008;54:517–527. - PubMed
-
- Hricak H, Wang L, Wei DC, Coakley FV, Akin O, Reuter VE, et al. The role of preoperative endorectal magnetic resonance imaging in the decision regarding whether to preserve or resect neurovascular bundles during radical retropubic prostatectomy. Cancer. 2004;100:2655–2663. - PubMed
-
- Liao S, Gao Y, Shi Y, Yousuf A, Karademir I, Oto A, et al. Automatic prostate MR image segmentation with sparse label propagation and domain-specific manifold regularization. In: Gee J, Joshi S, Pohl K, Wells W, Zöllei L, editors. Information Processing in Medical Imaging. Vol. 7917. Springer; Berlin Heidelberg: 2013. pp. 511–523. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

